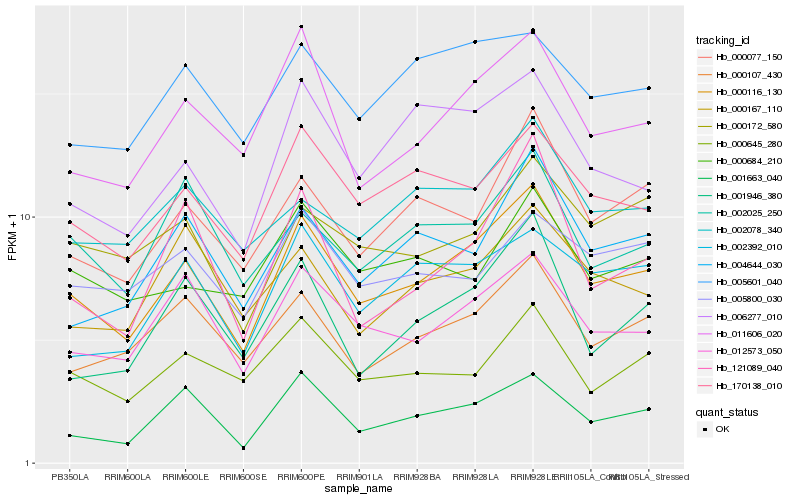
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_130 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 2 |
Hb_012573_050 |
0.0877497272 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 3 |
Hb_011606_020 |
0.0885162541 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 4 |
Hb_170138_010 |
0.0889389299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000077_150 |
0.0966074927 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 6 |
Hb_000107_430 |
0.0966216713 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 7 |
Hb_001663_040 |
0.0979210508 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 8 |
Hb_006277_010 |
0.1050741405 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 9 |
Hb_000645_280 |
0.1058445341 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 10 |
Hb_004644_030 |
0.1060269609 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 11 |
Hb_002392_010 |
0.1090820639 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 12 |
Hb_005601_040 |
0.1097374409 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 13 |
Hb_001946_380 |
0.1106695624 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 14 |
Hb_000172_580 |
0.1111980544 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002078_340 |
0.1112379668 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002025_250 |
0.1130546185 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 17 |
Hb_121089_040 |
0.1146926437 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 18 |
Hb_000167_110 |
0.1160167864 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000684_210 |
0.1162315716 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 20 |
Hb_005800_030 |
0.1166478974 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |