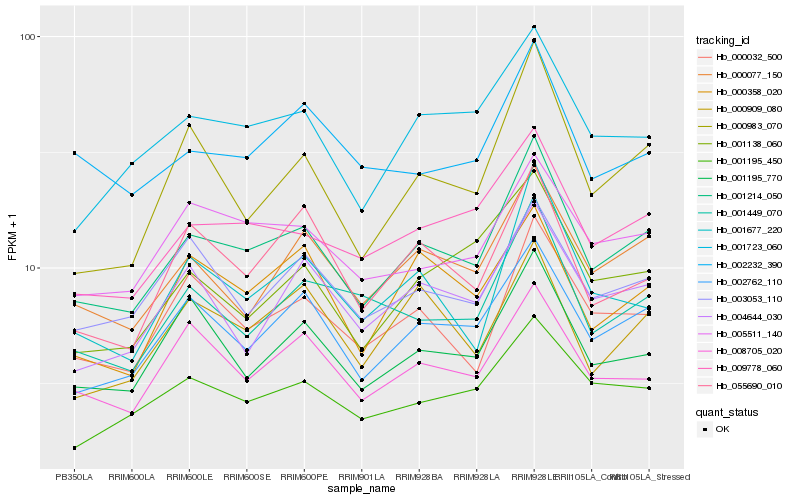
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001214_050 |
0.0 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002762_110 |
0.0706848253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009778_060 |
0.0786351591 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 4 |
Hb_000077_150 |
0.0789506958 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 5 |
Hb_001723_060 |
0.0869576296 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 6 |
Hb_000032_500 |
0.0876974281 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 7 |
Hb_001195_770 |
0.0913886781 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 8 |
Hb_005511_140 |
0.0915704079 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 9 |
Hb_001195_450 |
0.092084121 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 10 |
Hb_008705_020 |
0.0943331398 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_000983_070 |
0.0946456757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003053_110 |
0.0966195876 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001449_070 |
0.0974994895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001677_220 |
0.0975639247 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 15 |
Hb_004644_030 |
0.0982605348 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 16 |
Hb_055690_010 |
0.0993076288 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002232_390 |
0.1005783555 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 18 |
Hb_000358_020 |
0.1015503201 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 19 |
Hb_000909_080 |
0.1018907984 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 20 |
Hb_001138_060 |
0.1020516323 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |