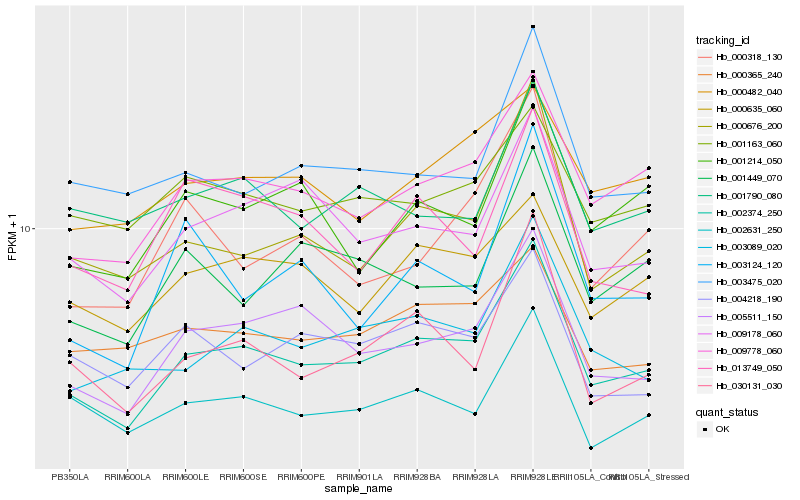
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009778_060 |
0.0851663499 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 3 |
Hb_005511_150 |
0.0870685681 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic isoform X5 [Populus euphratica] |
| 4 |
Hb_000676_200 |
0.0934090013 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 5 |
Hb_003089_020 |
0.096821948 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g18940, chloroplastic [Jatropha curcas] |
| 6 |
Hb_030131_030 |
0.099399631 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g42310, mitochondrial [Jatropha curcas] |
| 7 |
Hb_009178_060 |
0.1030773135 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002631_250 |
0.1078824907 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g20730 [Jatropha curcas] |
| 9 |
Hb_001214_050 |
0.1119598216 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003475_020 |
0.1129892318 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 11 |
Hb_001449_070 |
0.1158172759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004218_190 |
0.1181700093 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000318_130 |
0.1201178368 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000635_060 |
0.1204601652 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 15 |
Hb_000365_240 |
0.1234114122 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 16 |
Hb_013749_050 |
0.1239230703 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003124_120 |
0.1241788953 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 18 |
Hb_001790_080 |
0.1254927188 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 19 |
Hb_000482_040 |
0.1268656861 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 20 |
Hb_001163_060 |
0.128951876 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |