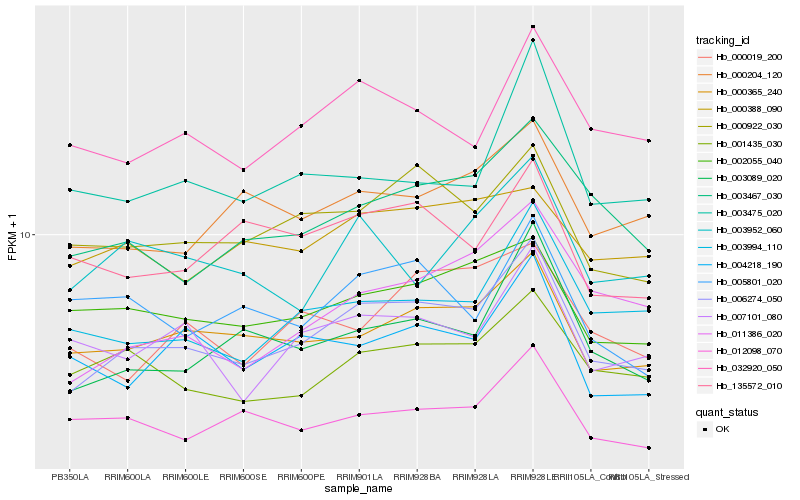
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006274_050 |
0.0 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 2 |
Hb_000365_240 |
0.0959332994 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 3 |
Hb_003089_020 |
0.1002106788 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g18940, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000922_030 |
0.102684052 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003467_030 |
0.1051259113 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 6 |
Hb_003994_110 |
0.1093416398 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 7 |
Hb_001435_030 |
0.1097774695 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 8 |
Hb_011386_020 |
0.1115997137 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_032920_050 |
0.11173677 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002055_040 |
0.1125194461 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 11 |
Hb_000204_120 |
0.1142216154 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 12 |
Hb_007101_080 |
0.1153794522 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 13 |
Hb_004218_190 |
0.1197168868 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 14 |
Hb_012098_070 |
0.1229356997 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 15 |
Hb_003952_060 |
0.1233826112 |
- |
- |
hypothetical protein POPTR_0016s11000g [Populus trichocarpa] |
| 16 |
Hb_135572_010 |
0.1244073464 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 17 |
Hb_000019_200 |
0.1260373264 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_003475_020 |
0.1277815544 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 19 |
Hb_000388_090 |
0.1302347867 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_005801_020 |
0.1305693623 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |