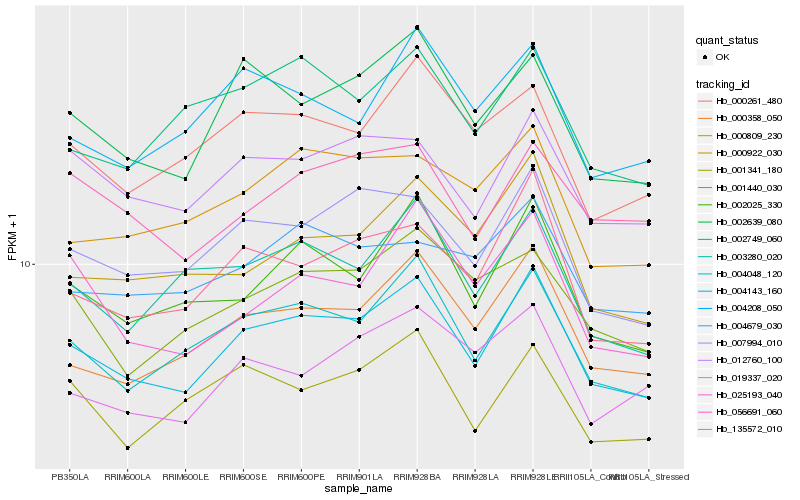
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004143_160 |
0.0 |
- |
- |
SAB, putative [Ricinus communis] |
| 2 |
Hb_004048_120 |
0.0495403691 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000358_050 |
0.0561117567 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 4 |
Hb_007994_010 |
0.0601611014 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002025_330 |
0.06031412 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 6 |
Hb_135572_010 |
0.0607850585 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 7 |
Hb_002639_080 |
0.0642042086 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_012760_100 |
0.0696192944 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 9 |
Hb_003280_020 |
0.0700126482 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000261_480 |
0.0735787915 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 11 |
Hb_000922_030 |
0.0746878778 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001341_180 |
0.0752495313 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 13 |
Hb_001440_030 |
0.0764240115 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 14 |
Hb_004208_050 |
0.0769185855 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 15 |
Hb_000809_230 |
0.0770048222 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 16 |
Hb_025193_040 |
0.0783259102 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 17 |
Hb_002749_060 |
0.079095236 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 18 |
Hb_019337_020 |
0.0792388636 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 19 |
Hb_056691_060 |
0.0799327353 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004679_030 |
0.080265123 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |