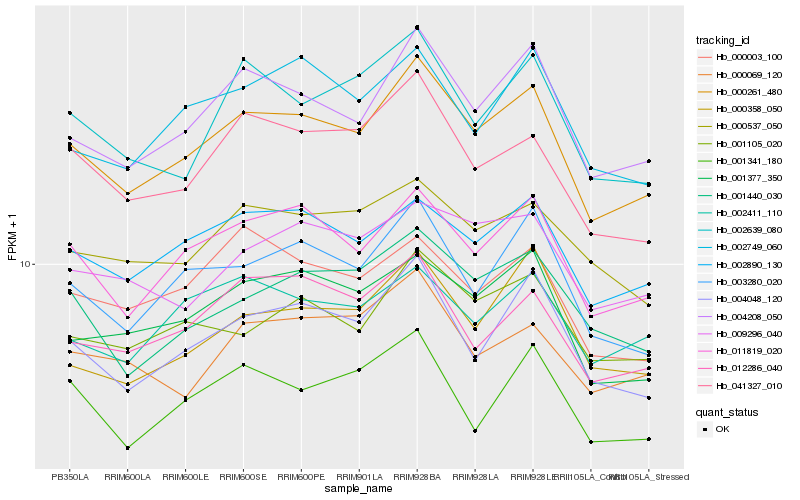
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_480 |
0.0 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 2 |
Hb_004208_050 |
0.0390134628 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 3 |
Hb_011819_020 |
0.0447091431 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004048_120 |
0.053196115 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 5 |
Hb_002890_130 |
0.0537087732 |
- |
- |
tip120, putative [Ricinus communis] |
| 6 |
Hb_003280_020 |
0.055760205 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001377_350 |
0.0572877181 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 8 |
Hb_041327_010 |
0.0582591124 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 9 |
Hb_001105_020 |
0.0620287883 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 10 |
Hb_001440_030 |
0.0642534499 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 11 |
Hb_012286_040 |
0.0647723093 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 12 |
Hb_002639_080 |
0.0660441977 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002749_060 |
0.0671409326 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 14 |
Hb_000537_050 |
0.0673702616 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 15 |
Hb_002411_110 |
0.0678636485 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 16 |
Hb_000358_050 |
0.0680689856 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 17 |
Hb_000003_100 |
0.0690224997 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 18 |
Hb_009296_040 |
0.0690951847 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 19 |
Hb_001341_180 |
0.0697498057 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 20 |
Hb_000069_120 |
0.0721731951 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |