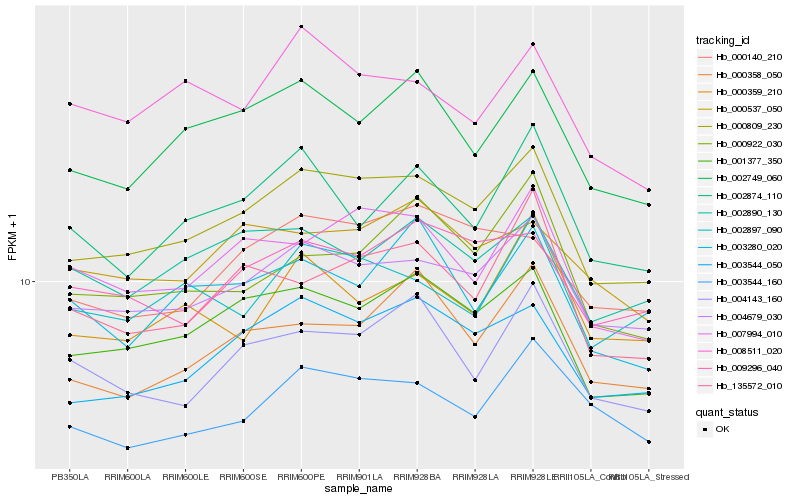
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004679_030 |
0.0417933499 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 3 |
Hb_001377_350 |
0.0473420006 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 4 |
Hb_003544_050 |
0.049870718 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002749_060 |
0.0634198556 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 6 |
Hb_007994_010 |
0.0659594158 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009296_040 |
0.0718929561 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 8 |
Hb_000358_050 |
0.073406687 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 9 |
Hb_000140_210 |
0.0747806766 |
- |
- |
sec10, putative [Ricinus communis] |
| 10 |
Hb_135572_010 |
0.0768088672 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 11 |
Hb_002890_130 |
0.076838457 |
- |
- |
tip120, putative [Ricinus communis] |
| 12 |
Hb_004143_160 |
0.0770048222 |
- |
- |
SAB, putative [Ricinus communis] |
| 13 |
Hb_003544_160 |
0.0782390019 |
- |
- |
- |
| 14 |
Hb_002897_090 |
0.0793230883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000537_050 |
0.0797401508 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 16 |
Hb_000922_030 |
0.0798519028 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002874_110 |
0.0805920893 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 18 |
Hb_000359_210 |
0.0831824297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003280_020 |
0.0834785622 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008511_020 |
0.0838494078 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |