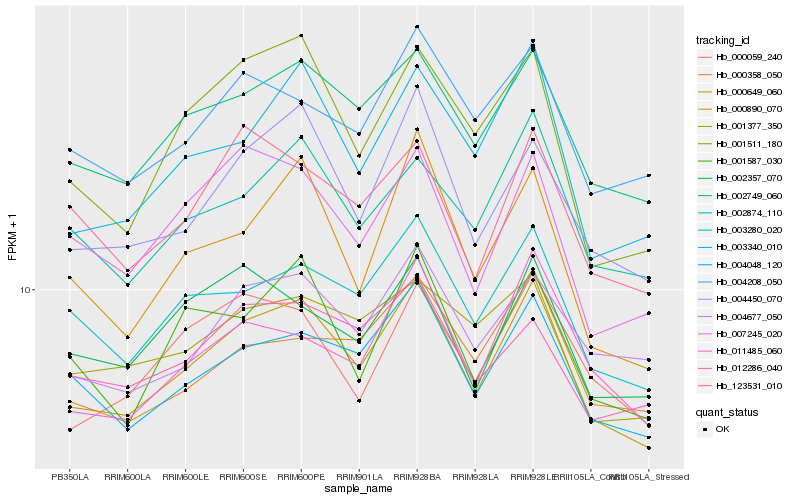
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_070 |
0.0 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002749_060 |
0.0484875853 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 3 |
Hb_004450_070 |
0.0583407298 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 4 |
Hb_004048_120 |
0.059604281 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 5 |
Hb_003280_020 |
0.0612921999 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003340_010 |
0.0665644263 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 7 |
Hb_002874_110 |
0.0677489741 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 8 |
Hb_001587_030 |
0.071088658 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 9 |
Hb_004677_050 |
0.0717673134 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000649_060 |
0.0720477877 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 11 |
Hb_011485_060 |
0.0728657278 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 12 |
Hb_000059_240 |
0.0734048258 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 13 |
Hb_004208_050 |
0.0734550998 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 14 |
Hb_001511_180 |
0.0743691949 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 15 |
Hb_007245_020 |
0.0752426115 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 16 |
Hb_002357_070 |
0.0754513107 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 17 |
Hb_001377_350 |
0.0758796729 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 18 |
Hb_123531_010 |
0.0764616385 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 19 |
Hb_012286_040 |
0.0771306682 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 20 |
Hb_000358_050 |
0.0772611502 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |