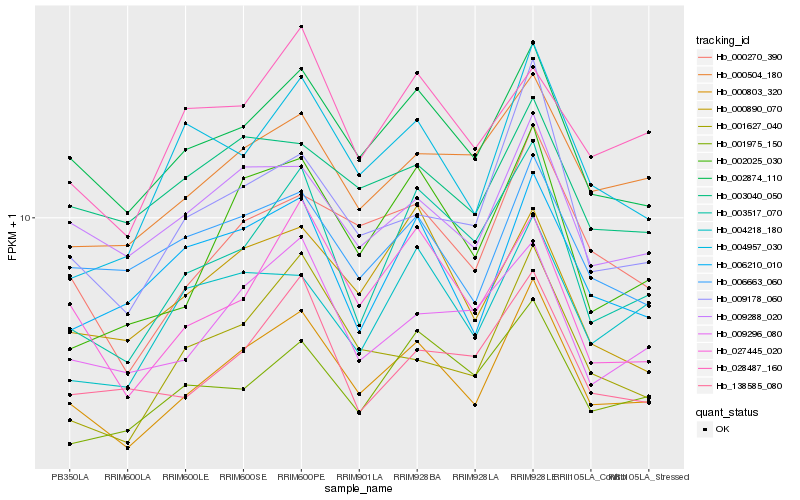
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_320 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 2 |
Hb_002874_110 |
0.0575524159 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 3 |
Hb_000270_390 |
0.0663454329 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 4 |
Hb_009288_020 |
0.0757696089 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 5 |
Hb_000504_180 |
0.0814095141 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_009178_060 |
0.0821237731 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_006663_060 |
0.0837656398 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 8 |
Hb_009296_080 |
0.0838717204 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003517_070 |
0.0855946302 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 10 |
Hb_027445_020 |
0.0871462571 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 11 |
Hb_006210_010 |
0.0882083622 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 12 |
Hb_001627_040 |
0.0885601578 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003040_050 |
0.08858239 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 14 |
Hb_002025_030 |
0.0889333437 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 15 |
Hb_138585_080 |
0.0891480708 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 16 |
Hb_001975_150 |
0.09049559 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 17 |
Hb_004957_030 |
0.0906466795 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 18 |
Hb_000890_070 |
0.0921550474 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004218_180 |
0.0922595484 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 20 |
Hb_028487_160 |
0.0923402807 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |