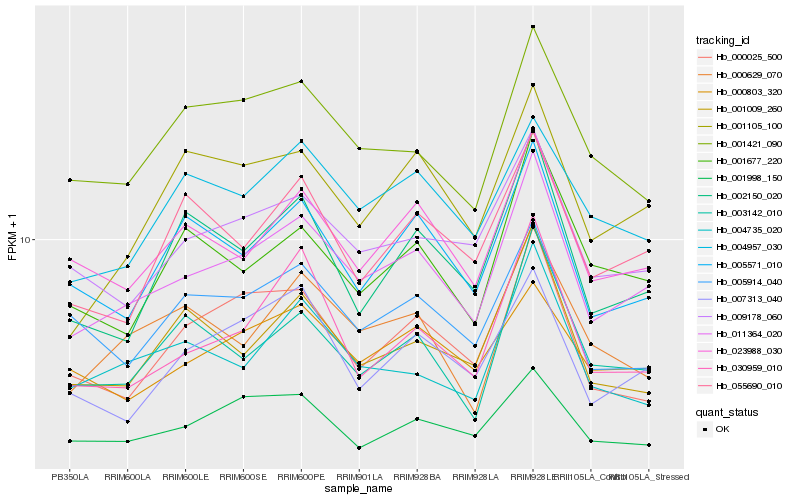
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011364_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 2 |
Hb_023988_030 |
0.09256116 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 3 |
Hb_009178_060 |
0.09373791 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_005571_010 |
0.1038512491 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 5 |
Hb_001421_090 |
0.1074138601 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 6 |
Hb_001105_100 |
0.1088089827 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 7 |
Hb_030959_010 |
0.1104092704 |
- |
- |
cullin, putative [Ricinus communis] |
| 8 |
Hb_002150_020 |
0.1114257204 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_001677_220 |
0.1118058342 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 10 |
Hb_003142_010 |
0.1118970545 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 11 |
Hb_001009_260 |
0.1127173939 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004735_020 |
0.1131405833 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 13 |
Hb_000629_070 |
0.1134414201 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 14 |
Hb_007313_040 |
0.1134628466 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 15 |
Hb_004957_030 |
0.1136869077 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 16 |
Hb_001998_150 |
0.1141725541 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 17 |
Hb_000803_320 |
0.1143439612 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 18 |
Hb_000025_500 |
0.1143787323 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 19 |
Hb_055690_010 |
0.1143815772 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005914_040 |
0.1162372301 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |