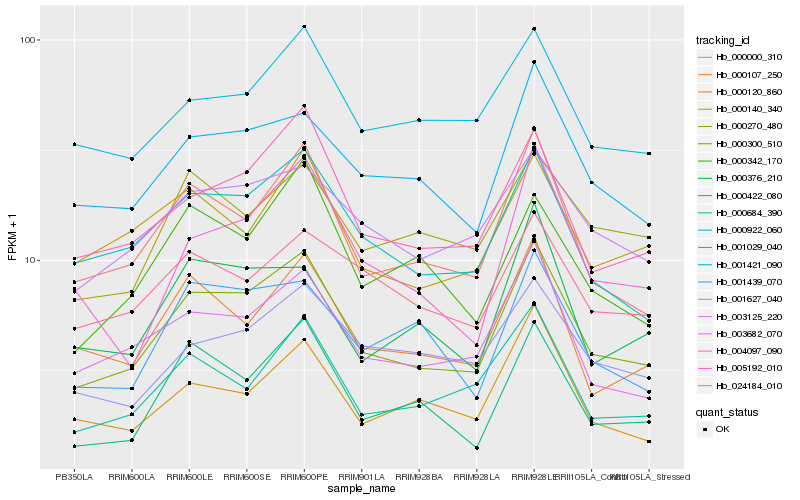
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_510 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 2 |
Hb_000000_310 |
0.0773687224 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 3 |
Hb_005192_010 |
0.085942476 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_000922_060 |
0.0885941183 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 5 |
Hb_003682_070 |
0.0886552013 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001421_090 |
0.0960492782 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 7 |
Hb_000684_390 |
0.1024165207 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 8 |
Hb_000107_250 |
0.1031174795 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 9 |
Hb_003125_220 |
0.1047372512 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 10 |
Hb_004097_090 |
0.1052667816 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 11 |
Hb_001439_070 |
0.1102744533 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001029_040 |
0.1103804952 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 13 |
Hb_000422_080 |
0.1105458687 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 14 |
Hb_000342_170 |
0.1111379365 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 15 |
Hb_001627_040 |
0.1118436632 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000140_340 |
0.1142451633 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 17 |
Hb_000376_210 |
0.1143558572 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 18 |
Hb_024184_010 |
0.1164786651 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_000120_860 |
0.1172613466 |
- |
- |
nucellin, putative [Ricinus communis] |
| 20 |
Hb_000270_480 |
0.1173810643 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |