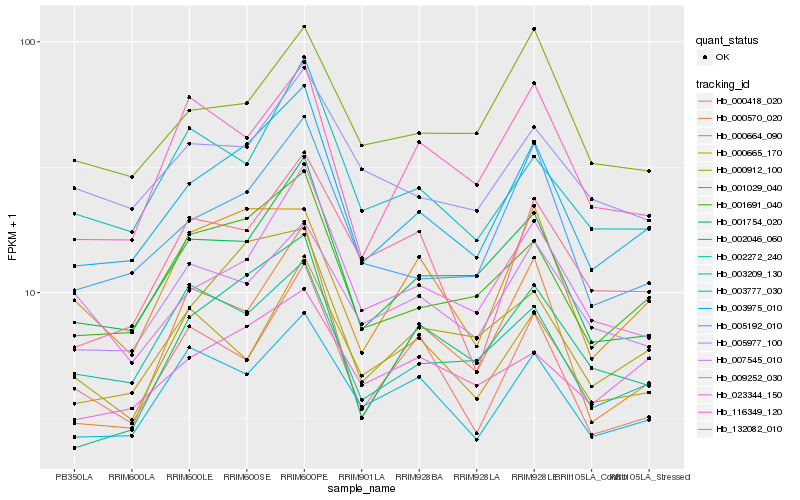
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003975_010 |
0.0 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 2 |
Hb_001691_040 |
0.066843507 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 3 |
Hb_005192_010 |
0.0799267623 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_001754_020 |
0.0803126251 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 5 |
Hb_000912_100 |
0.0906771909 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 6 |
Hb_002046_060 |
0.0985807268 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 7 |
Hb_009252_030 |
0.0996030452 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 8 |
Hb_003777_030 |
0.1011601037 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 9 |
Hb_003209_130 |
0.1047084137 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_007545_010 |
0.1059348077 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 11 |
Hb_002272_240 |
0.1074427357 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_023344_150 |
0.1092097806 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 13 |
Hb_000570_020 |
0.111279564 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 14 |
Hb_001029_040 |
0.1118708745 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 15 |
Hb_116349_120 |
0.1127896762 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_000665_170 |
0.1137226027 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 17 |
Hb_005977_100 |
0.1140720732 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 18 |
Hb_000664_090 |
0.1143151267 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 19 |
Hb_132082_010 |
0.1146502921 |
- |
- |
hypothetical protein VITISV_030895 [Vitis vinifera] |
| 20 |
Hb_000418_020 |
0.1150204072 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |