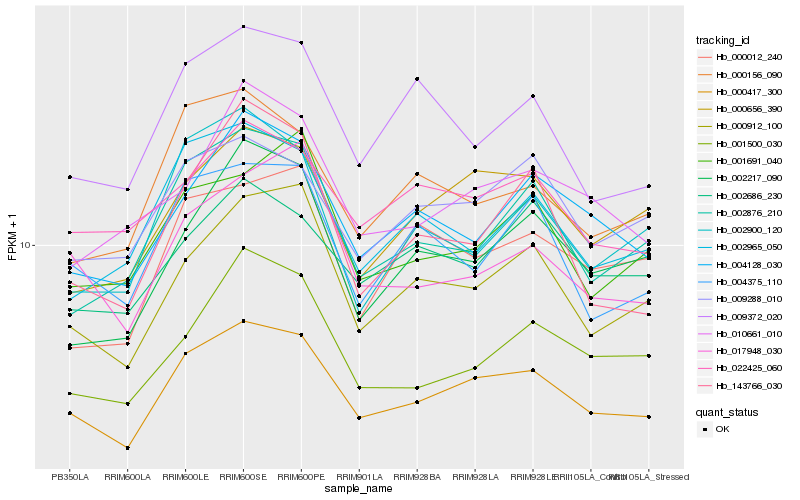
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_300 |
0.0 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 2 |
Hb_000912_100 |
0.0873319245 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 3 |
Hb_000656_390 |
0.0987482961 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002876_210 |
0.1008781083 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 5 |
Hb_004128_030 |
0.1010115186 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004375_110 |
0.1059629651 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 7 |
Hb_009288_010 |
0.108189987 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_002217_090 |
0.109281423 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 9 |
Hb_000156_090 |
0.110913252 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009372_020 |
0.1110393765 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 11 |
Hb_002965_050 |
0.1112119301 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 12 |
Hb_017948_030 |
0.1134989874 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 13 |
Hb_022425_060 |
0.1138987691 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_001500_030 |
0.114781538 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001691_040 |
0.1157209125 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000012_240 |
0.1170222778 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002900_120 |
0.1176970708 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 18 |
Hb_010661_010 |
0.1185838477 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 19 |
Hb_143766_030 |
0.1187282208 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_002686_230 |
0.1197169115 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |