| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
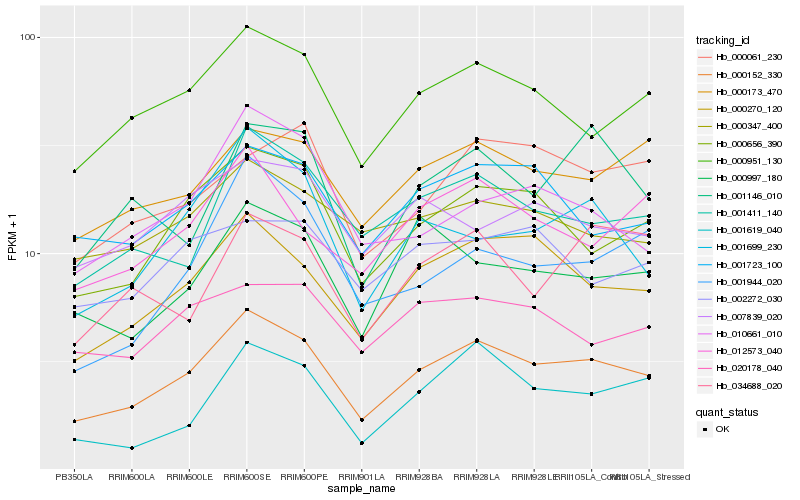
Hb_000152_330 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 2 |
Hb_000270_120 |
0.0870822163 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 3 |
Hb_000656_390 |
0.0891103656 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000951_130 |
0.0926120071 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 5 |
Hb_001411_140 |
0.1046870866 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 6 |
Hb_000173_470 |
0.1100328198 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_001723_100 |
0.1131591117 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001699_230 |
0.1158839417 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 9 |
Hb_012573_040 |
0.1160213557 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 10 |
Hb_000347_400 |
0.1214859393 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000997_180 |
0.1228637362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001146_010 |
0.1241557448 |
- |
- |
cystathionine gamma-synthase, putative [Ricinus communis] |
| 13 |
Hb_020178_040 |
0.1257932534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010661_010 |
0.1260625119 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 15 |
Hb_001944_020 |
0.1272179254 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 16 |
Hb_000061_230 |
0.1287024867 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_007839_020 |
0.1289723113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001619_040 |
0.1301612287 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 19 |
Hb_034688_020 |
0.1315471706 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 20 |
Hb_002272_030 |
0.1318373835 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |