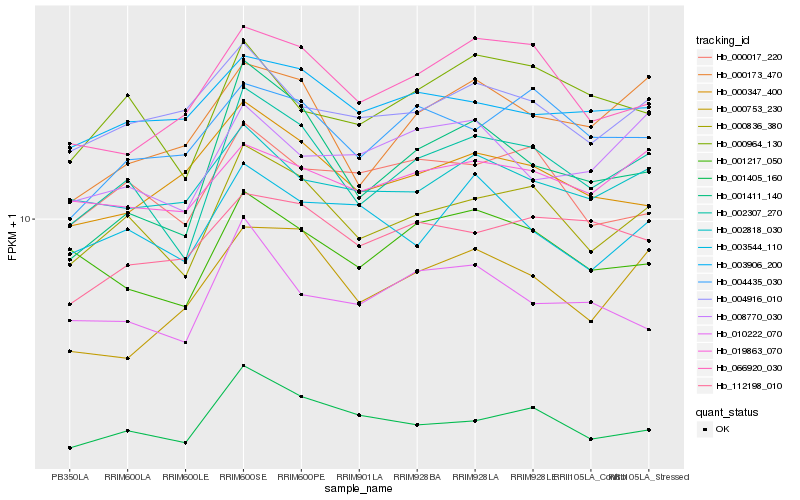
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002307_270 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 2 |
Hb_000836_380 |
0.0443763641 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 3 |
Hb_001411_140 |
0.0782738588 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 4 |
Hb_000964_130 |
0.0896961923 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 5 |
Hb_001405_160 |
0.0934631726 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 6 |
Hb_001217_050 |
0.0934668666 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 7 |
Hb_000173_470 |
0.0948911964 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_066920_030 |
0.1013531372 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 9 |
Hb_004435_030 |
0.103967315 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 10 |
Hb_010222_070 |
0.1041367878 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000017_220 |
0.1049422345 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 12 |
Hb_019863_070 |
0.1063833373 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_112198_010 |
0.1066009053 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 14 |
Hb_002818_030 |
0.1067282746 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 15 |
Hb_003906_200 |
0.1086630298 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 16 |
Hb_003544_110 |
0.1096969607 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_008770_030 |
0.1099595839 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290-like [Jatropha curcas] |
| 18 |
Hb_004916_010 |
0.1104737288 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 19 |
Hb_000753_230 |
0.1106476037 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000347_400 |
0.1115641038 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |