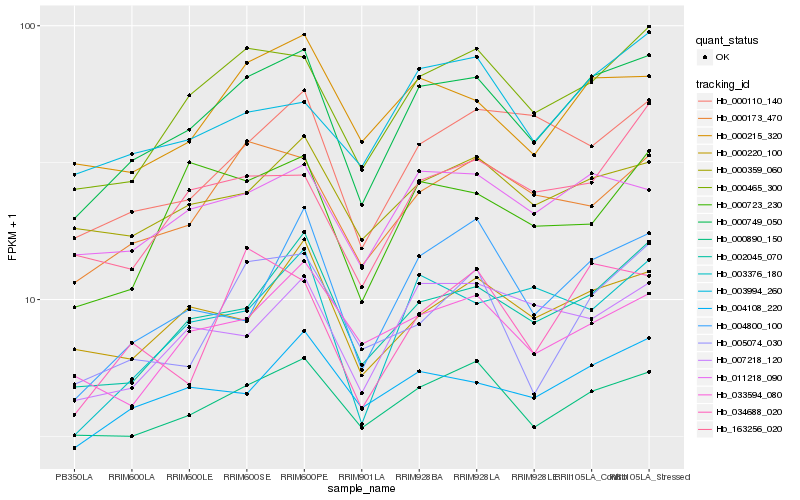
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_050 |
0.0 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 2 |
Hb_000465_300 |
0.0645803821 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 3 |
Hb_002045_070 |
0.0756615071 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 4 |
Hb_000173_470 |
0.0823000315 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_000890_150 |
0.0826871502 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Topors isoform X1 [Jatropha curcas] |
| 6 |
Hb_000215_320 |
0.0838728904 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 7 |
Hb_011218_090 |
0.0841368246 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 8 |
Hb_004108_220 |
0.0902157122 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000359_060 |
0.0910013046 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 10 |
Hb_000220_100 |
0.0924189459 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 [Jatropha curcas] |
| 11 |
Hb_007218_120 |
0.0953002064 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000110_140 |
0.0954859407 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 13 |
Hb_034688_020 |
0.0962075918 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 14 |
Hb_004800_100 |
0.0985457775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003376_180 |
0.0987858139 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 16 |
Hb_033594_080 |
0.0999130835 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 17 |
Hb_003994_260 |
0.100190614 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 18 |
Hb_005074_030 |
0.1004968393 |
- |
- |
- |
| 19 |
Hb_163256_020 |
0.100928263 |
- |
- |
fructokinase [Manihot esculenta] |
| 20 |
Hb_000723_230 |
0.1018733141 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |