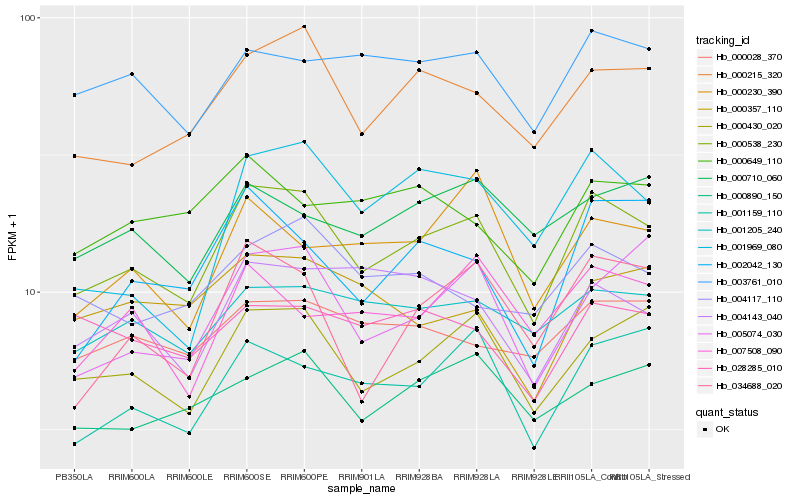
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_230 |
0.0 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000430_020 |
0.0759291265 |
- |
- |
- |
| 3 |
Hb_000215_320 |
0.0808032008 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 4 |
Hb_001159_110 |
0.0962448523 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 5 |
Hb_005074_030 |
0.0979355283 |
- |
- |
- |
| 6 |
Hb_001205_240 |
0.0991919962 |
- |
- |
PREDICTED: uncharacterized protein LOC105645528 [Jatropha curcas] |
| 7 |
Hb_003761_010 |
0.099555946 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_034688_020 |
0.1008739595 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 9 |
Hb_000890_150 |
0.1019285181 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Topors isoform X1 [Jatropha curcas] |
| 10 |
Hb_002042_130 |
0.1027732631 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 11 |
Hb_000028_370 |
0.1057043498 |
- |
- |
Guanine nucleotide-binding protein alpha-2 subunit [Glycine soja] |
| 12 |
Hb_004143_040 |
0.1057438374 |
- |
- |
argininosuccinate lyase, putative [Ricinus communis] |
| 13 |
Hb_004117_110 |
0.1065722767 |
- |
- |
hypothetical protein POPTR_0016s14420g [Populus trichocarpa] |
| 14 |
Hb_001969_080 |
0.107097292 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 15 |
Hb_000710_060 |
0.1073966401 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105645554 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000230_390 |
0.1092353805 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 17 |
Hb_007508_090 |
0.1110600094 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 18 |
Hb_028285_010 |
0.1121544948 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 19 |
Hb_000649_110 |
0.1127258167 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 20 |
Hb_000357_110 |
0.1135555559 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |