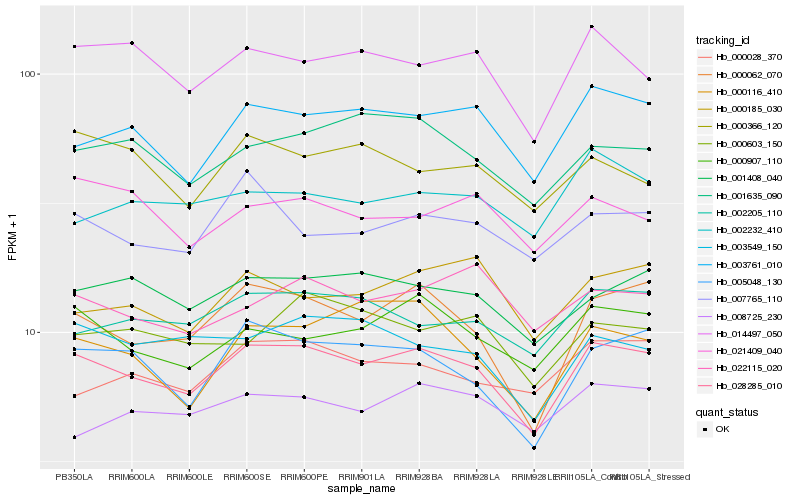
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028285_010 |
0.0 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 2 |
Hb_000062_070 |
0.0612152104 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 3 |
Hb_005048_130 |
0.0651612253 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 4 |
Hb_000907_110 |
0.0658099289 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 5 |
Hb_003761_010 |
0.0665536712 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001408_040 |
0.0679362233 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 7 |
Hb_014497_050 |
0.0691706937 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_08021 [Jatropha curcas] |
| 8 |
Hb_002205_110 |
0.0717823471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001635_090 |
0.073471886 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 10 |
Hb_003549_150 |
0.075042441 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 11 |
Hb_007765_110 |
0.0763772292 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 12 |
Hb_000028_370 |
0.0784351346 |
- |
- |
Guanine nucleotide-binding protein alpha-2 subunit [Glycine soja] |
| 13 |
Hb_000116_410 |
0.0797823327 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 14 |
Hb_000603_150 |
0.079792511 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 15 |
Hb_000185_030 |
0.0807611074 |
- |
- |
PREDICTED: arginine-glutamic acid dipeptide repeats protein-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_022115_020 |
0.0809119681 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002232_410 |
0.0810584343 |
- |
- |
PREDICTED: L-galactose dehydrogenase [Jatropha curcas] |
| 18 |
Hb_021409_040 |
0.0820831804 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 19 |
Hb_008725_230 |
0.0822215221 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 20 |
Hb_000366_120 |
0.082318879 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |