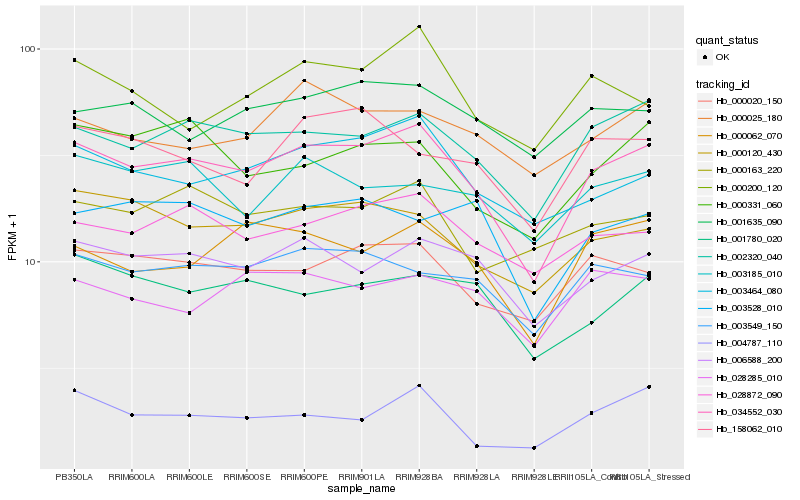
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034552_030 |
0.0 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_002320_040 |
0.0718336538 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 3 |
Hb_006588_200 |
0.0831063875 |
- |
- |
3-beta-hydroxy-delta5-steroid dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000020_150 |
0.085879968 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 5 |
Hb_003464_080 |
0.085900331 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 6 |
Hb_028872_090 |
0.0877144966 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 7 |
Hb_000120_430 |
0.0902643432 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 8 |
Hb_003549_150 |
0.0928458716 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 9 |
Hb_000062_070 |
0.0931386275 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 10 |
Hb_001780_020 |
0.100695379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000331_060 |
0.1013274605 |
- |
- |
PREDICTED: 60S ribosomal protein L17-2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000025_180 |
0.1030094177 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 13 |
Hb_000163_220 |
0.1036693045 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 14 |
Hb_001635_090 |
0.1039552384 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 15 |
Hb_004787_110 |
0.1043291426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_028285_010 |
0.1045946409 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 17 |
Hb_158062_010 |
0.1046528412 |
- |
- |
PREDICTED: uncharacterized protein LOC105634413 [Jatropha curcas] |
| 18 |
Hb_003528_010 |
0.104966634 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 3R-1 [Jatropha curcas] |
| 19 |
Hb_003185_010 |
0.1053465274 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |
| 20 |
Hb_000200_120 |
0.105387138 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |