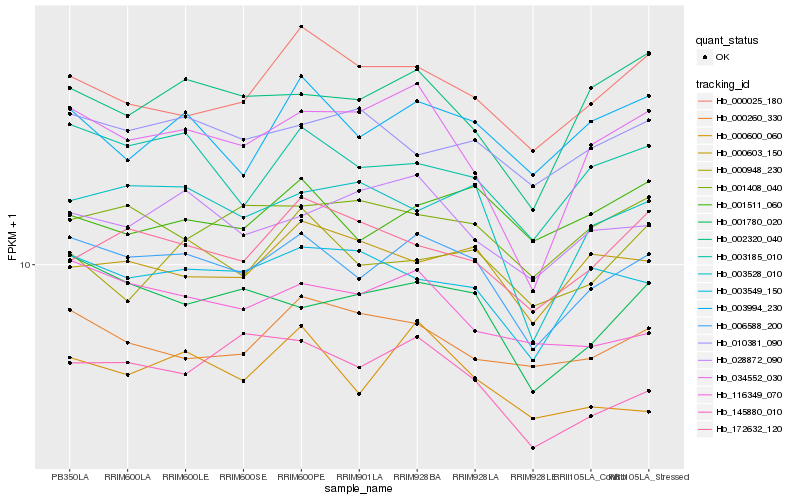
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_200 |
0.0 |
- |
- |
3-beta-hydroxy-delta5-steroid dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_003185_010 |
0.0677012756 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |
| 3 |
Hb_001780_020 |
0.0740353225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003549_150 |
0.0805338231 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 5 |
Hb_003528_010 |
0.0829886078 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 3R-1 [Jatropha curcas] |
| 6 |
Hb_034552_030 |
0.0831063875 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_003994_230 |
0.0831492032 |
- |
- |
7-dehydrocholesterol reductase, putative [Ricinus communis] |
| 8 |
Hb_028872_090 |
0.0860353018 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 9 |
Hb_116349_070 |
0.0861444423 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 10 |
Hb_000025_180 |
0.0862835831 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 11 |
Hb_001408_040 |
0.0863403119 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002320_040 |
0.0874133672 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 13 |
Hb_000603_150 |
0.0888152562 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 14 |
Hb_010381_090 |
0.0895058998 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 15 |
Hb_000948_230 |
0.0909607676 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_000600_060 |
0.0916912207 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000260_330 |
0.0917010303 |
- |
- |
PREDICTED: uncharacterized protein LOC103438625 [Malus domestica] |
| 18 |
Hb_172632_120 |
0.092071022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_145880_010 |
0.0921842065 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 20 |
Hb_001511_060 |
0.0933155558 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |