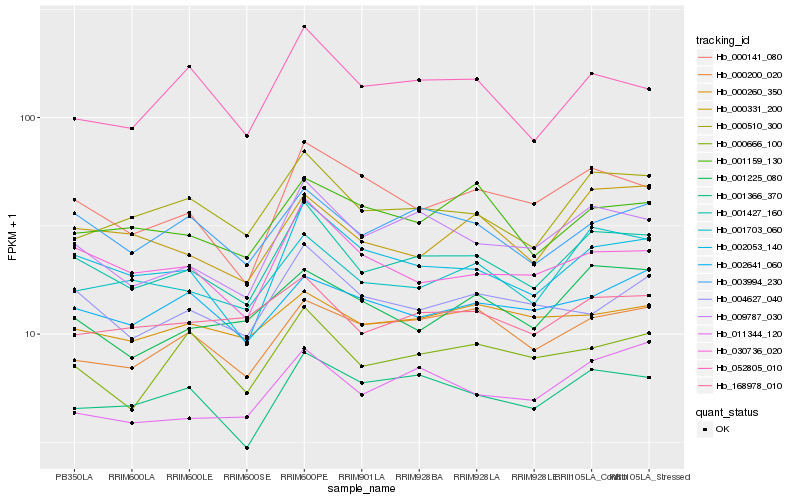
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_160 |
0.0 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_009787_030 |
0.0604666514 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 3 |
Hb_002053_140 |
0.063800746 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 4 |
Hb_000510_300 |
0.0656882424 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003994_230 |
0.0667740543 |
- |
- |
7-dehydrocholesterol reductase, putative [Ricinus communis] |
| 6 |
Hb_001366_370 |
0.06918872 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001703_060 |
0.0692710831 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 8 |
Hb_030736_020 |
0.0717734509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_168978_010 |
0.0746843053 |
- |
- |
PREDICTED: uncharacterized protein LOC105630751 [Jatropha curcas] |
| 10 |
Hb_000141_080 |
0.0774048113 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 11 |
Hb_004627_040 |
0.0781955068 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 12 |
Hb_000200_020 |
0.0803372658 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 13 |
Hb_000331_200 |
0.080750463 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 14 |
Hb_052805_010 |
0.0816661 |
- |
- |
hypothetical protein POPTR_0009s03650g [Populus trichocarpa] |
| 15 |
Hb_000666_100 |
0.0824902018 |
- |
- |
Actin-related protein 2/3 complex subunit 2 isoform 2 [Theobroma cacao] |
| 16 |
Hb_001159_130 |
0.084000693 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 17 |
Hb_002641_060 |
0.0848911899 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |
| 18 |
Hb_000260_350 |
0.0856355732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011344_120 |
0.0857413265 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 20 |
Hb_001225_080 |
0.0857911018 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8A [Populus euphratica] |