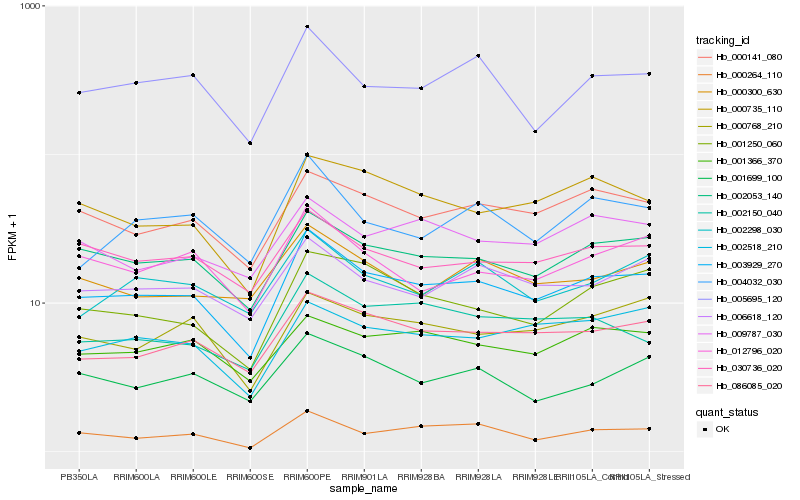
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_270 |
0.0 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002053_140 |
0.0681475936 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 3 |
Hb_086085_020 |
0.0794634478 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 4 |
Hb_000141_080 |
0.0802695686 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 5 |
Hb_000735_110 |
0.0901632765 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 6 |
Hb_030736_020 |
0.0914897456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006618_120 |
0.0916064635 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 8 |
Hb_001250_060 |
0.0920556083 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 9 |
Hb_000264_110 |
0.0930701142 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 13 [Jatropha curcas] |
| 10 |
Hb_005695_120 |
0.0932272809 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 11 |
Hb_012796_020 |
0.0953956271 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |
| 12 |
Hb_001366_370 |
0.096923198 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000768_210 |
0.0970429385 |
- |
- |
PREDICTED: uncharacterized protein LOC105632522 [Jatropha curcas] |
| 14 |
Hb_002518_210 |
0.0971551697 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |
| 15 |
Hb_001699_100 |
0.0996544272 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 16 |
Hb_002298_030 |
0.1007011116 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 17 |
Hb_002150_040 |
0.1008907704 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000300_630 |
0.1010818081 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 19 |
Hb_004032_030 |
0.1029389098 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 20 |
Hb_009787_030 |
0.1030823658 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |