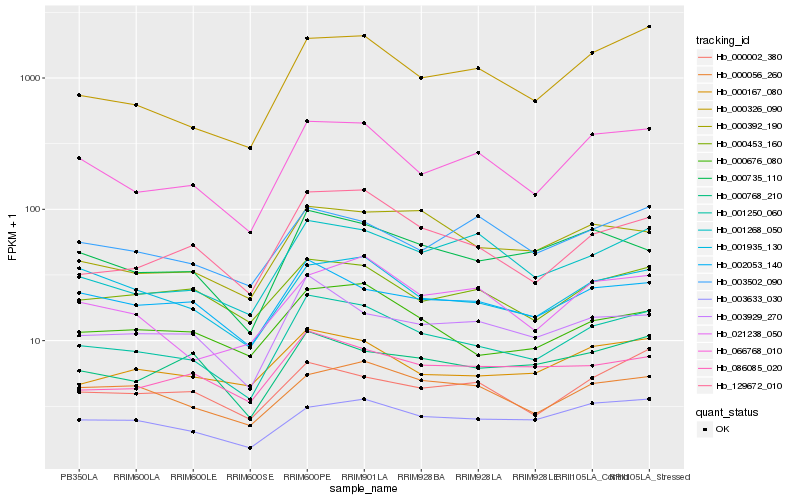
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001250_060 |
0.0 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 2 |
Hb_066768_010 |
0.0781508341 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_003929_270 |
0.0920556083 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_129672_010 |
0.0936736925 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 5 |
Hb_000735_110 |
0.0940494156 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 6 |
Hb_000326_090 |
0.0952079132 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 7 |
Hb_000167_080 |
0.0957406996 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 8 |
Hb_000676_080 |
0.0961358636 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 9 |
Hb_002053_140 |
0.0985133005 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 10 |
Hb_001268_050 |
0.0999834505 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 11 |
Hb_000453_160 |
0.1024090276 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 12 |
Hb_003633_030 |
0.1031659111 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 13 |
Hb_000392_190 |
0.1038137976 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 14 |
Hb_086085_020 |
0.1040636988 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 15 |
Hb_001935_130 |
0.1062065374 |
- |
- |
PREDICTED: uncharacterized protein LOC105643176 [Jatropha curcas] |
| 16 |
Hb_000056_260 |
0.107133936 |
- |
- |
hypothetical protein CICLE_v10033822mg [Citrus clementina] |
| 17 |
Hb_000768_210 |
0.1080111325 |
- |
- |
PREDICTED: uncharacterized protein LOC105632522 [Jatropha curcas] |
| 18 |
Hb_021238_050 |
0.110519452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003502_090 |
0.1114891665 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 20 |
Hb_000002_380 |
0.1119236353 |
- |
- |
PREDICTED: bet1-like SNARE 1-2 [Jatropha curcas] |