| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
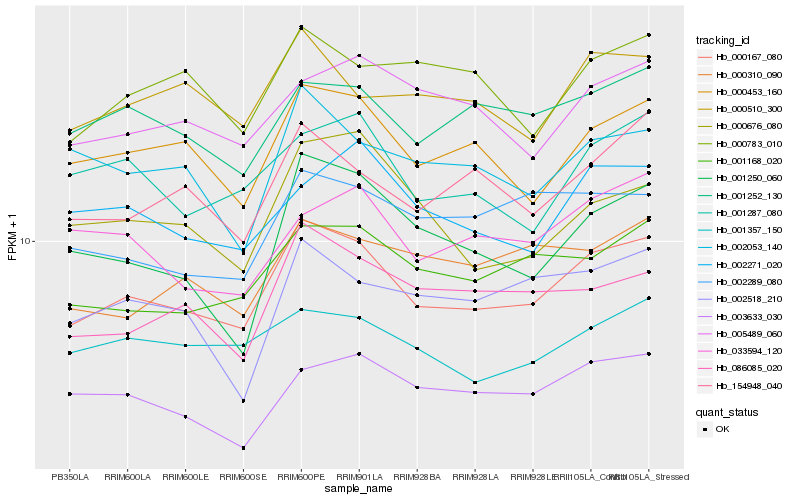
Hb_000167_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 2 |
Hb_000453_160 |
0.0770659255 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 3 |
Hb_001168_020 |
0.0863927356 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit gamma [Jatropha curcas] |
| 4 |
Hb_154948_040 |
0.0908686492 |
- |
- |
hypothetical protein B456_013G043800 [Gossypium raimondii] |
| 5 |
Hb_086085_020 |
0.0934864453 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 6 |
Hb_001252_130 |
0.0939714804 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 7 |
Hb_001287_080 |
0.0954254027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001250_060 |
0.0957406996 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 9 |
Hb_001357_150 |
0.0960871483 |
- |
- |
hypothetical protein POPTR_0011s08930g [Populus trichocarpa] |
| 10 |
Hb_000510_300 |
0.097483014 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_005489_060 |
0.1009987463 |
- |
- |
Anamorsin, putative [Ricinus communis] |
| 12 |
Hb_002271_020 |
0.1014344002 |
- |
- |
hypothetical protein EUGRSUZ_G02641 [Eucalyptus grandis] |
| 13 |
Hb_000676_080 |
0.1017811259 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 14 |
Hb_000310_090 |
0.102081585 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002053_140 |
0.1025106262 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 16 |
Hb_003633_030 |
0.1030088217 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 17 |
Hb_002289_080 |
0.1031416824 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |
| 18 |
Hb_000783_010 |
0.1032207506 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 19 |
Hb_002518_210 |
0.10399207 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |
| 20 |
Hb_033594_120 |
0.1058819183 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |