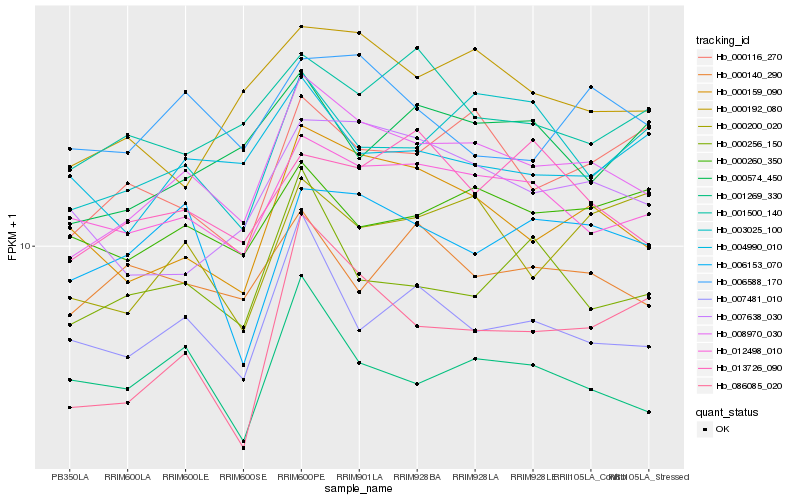
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008970_030 |
0.0 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 2 |
Hb_012498_010 |
0.0648953221 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 3 |
Hb_000159_090 |
0.0675583918 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 4 |
Hb_007481_010 |
0.0760524543 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 5 |
Hb_003025_100 |
0.0781153835 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000192_080 |
0.0797342756 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_013726_090 |
0.0803841687 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 8 |
Hb_001269_330 |
0.0804224227 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 9 |
Hb_000116_270 |
0.08047737 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |
| 10 |
Hb_000256_150 |
0.080648129 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 11 |
Hb_086085_020 |
0.0814880768 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 12 |
Hb_001500_140 |
0.0820503221 |
- |
- |
pelota, putative [Ricinus communis] |
| 13 |
Hb_007638_030 |
0.0826399201 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 14 |
Hb_006588_170 |
0.0835181635 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 15 |
Hb_000574_450 |
0.0839102638 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 16 |
Hb_006153_070 |
0.084128982 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000260_350 |
0.0843471905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000140_290 |
0.0844202723 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 59 kDa protein isoform X1 [Populus euphratica] |
| 19 |
Hb_000200_020 |
0.0847680256 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 20 |
Hb_004990_010 |
0.0852490269 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |