| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
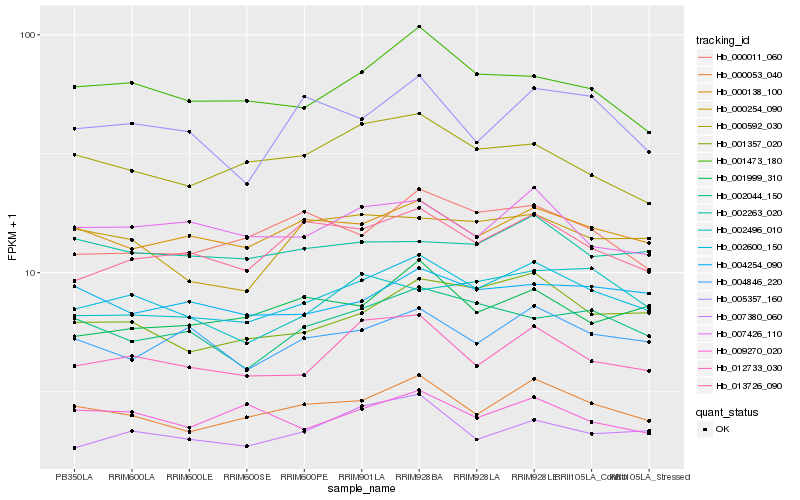
Hb_002600_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000053_040 |
0.0472786078 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 3 |
Hb_012733_030 |
0.0533061931 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 4 |
Hb_001357_020 |
0.0544271219 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000138_100 |
0.0601889428 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 6 |
Hb_013726_090 |
0.0607466017 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 7 |
Hb_001473_180 |
0.0638223715 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 8 |
Hb_007380_060 |
0.0664885718 |
- |
- |
hypothetical protein POPTR_0006s00280g [Populus trichocarpa] |
| 9 |
Hb_007426_110 |
0.0670609719 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 10 |
Hb_004846_220 |
0.0671297025 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000011_060 |
0.0697750019 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002044_150 |
0.0697810447 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 13 |
Hb_001999_310 |
0.0708789803 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 14 |
Hb_000592_030 |
0.0727960563 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 15 |
Hb_004254_090 |
0.0730159606 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 16 |
Hb_000254_090 |
0.0736968249 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 17 |
Hb_002263_020 |
0.0743252664 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 18 |
Hb_002496_010 |
0.074900069 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_005357_160 |
0.0757187041 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 20 |
Hb_009270_020 |
0.0761881373 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |