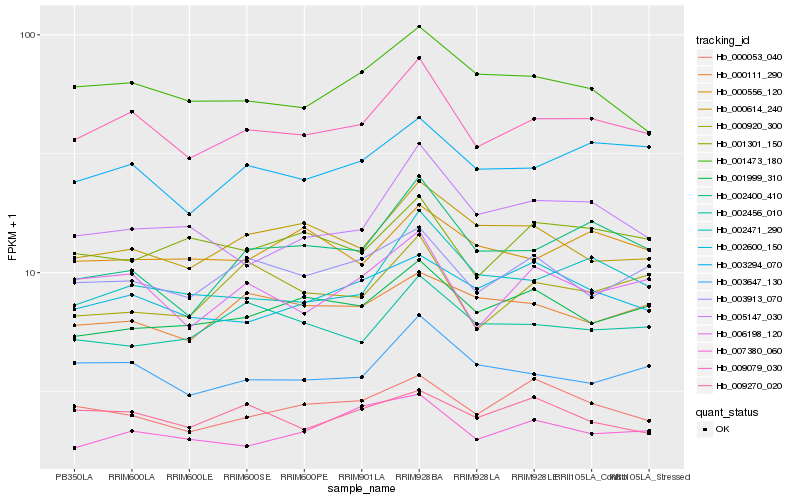
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009079_030 |
0.0 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 2 |
Hb_003647_130 |
0.0615648887 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 3 |
Hb_006198_120 |
0.0620870347 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 4 |
Hb_003294_070 |
0.0625650951 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 5 |
Hb_002471_290 |
0.0645611563 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_003913_070 |
0.0717001163 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 7 |
Hb_001473_180 |
0.072039138 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 8 |
Hb_001999_310 |
0.0766607132 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 9 |
Hb_007380_060 |
0.0769548652 |
- |
- |
hypothetical protein POPTR_0006s00280g [Populus trichocarpa] |
| 10 |
Hb_002400_410 |
0.0781150763 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 11 |
Hb_000053_040 |
0.0783588014 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 12 |
Hb_000614_240 |
0.0784033721 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 13 |
Hb_002600_150 |
0.0784873317 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005147_030 |
0.0803514994 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 15 |
Hb_001301_150 |
0.0809408171 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 16 |
Hb_002456_010 |
0.0812615962 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_000556_120 |
0.0829814821 |
- |
- |
Spastin, putative [Ricinus communis] |
| 18 |
Hb_000920_300 |
0.0830882138 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 19 |
Hb_009270_020 |
0.083235389 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 20 |
Hb_000111_290 |
0.0839552026 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |