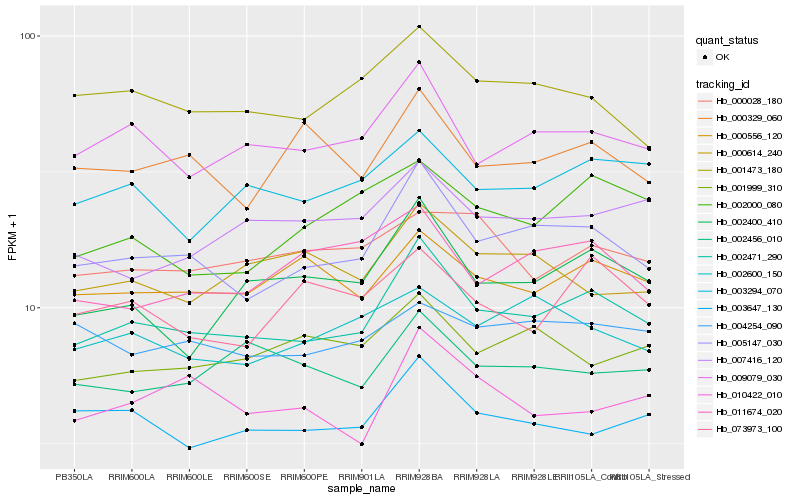
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002471_290 |
0.0 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_005147_030 |
0.0474325634 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 3 |
Hb_009079_030 |
0.0645611563 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 4 |
Hb_002400_410 |
0.0775186582 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 5 |
Hb_003294_070 |
0.0776933101 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 6 |
Hb_000556_120 |
0.0797896021 |
- |
- |
Spastin, putative [Ricinus communis] |
| 7 |
Hb_003647_130 |
0.0799556531 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 8 |
Hb_001473_180 |
0.080895311 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 9 |
Hb_002456_010 |
0.0882510566 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_000329_060 |
0.0895873876 |
- |
- |
chloroplast 5-enolpyruvylshikimate 3-phosphate synthase [Hevea brasiliensis] |
| 11 |
Hb_007416_120 |
0.0897147642 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 12 |
Hb_010422_010 |
0.0904445464 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 13 |
Hb_002600_150 |
0.0909354679 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002000_080 |
0.0911514587 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001999_310 |
0.091795652 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 16 |
Hb_004254_090 |
0.0919571019 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 17 |
Hb_073973_100 |
0.092112367 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 18 |
Hb_011674_020 |
0.092771136 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 19 |
Hb_000028_180 |
0.093758715 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000614_240 |
0.0941040335 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |