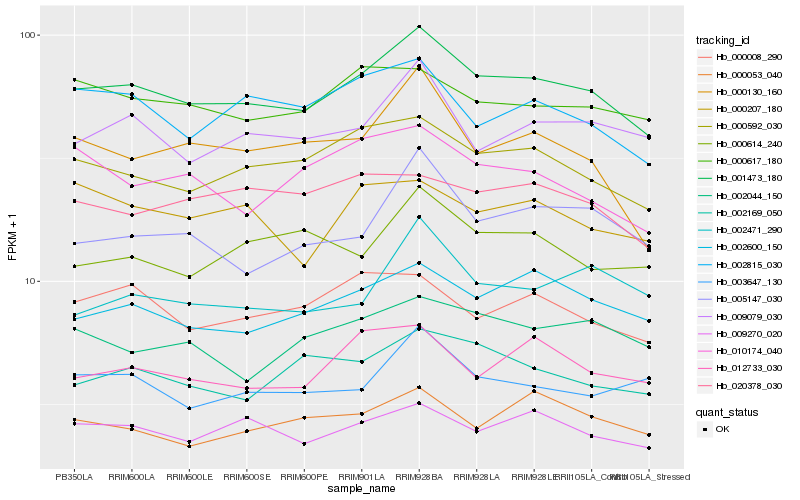
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001473_180 |
0.0 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 2 |
Hb_000592_030 |
0.0578019446 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 3 |
Hb_002600_150 |
0.0638223715 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009270_020 |
0.0656614491 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 5 |
Hb_009079_030 |
0.072039138 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 6 |
Hb_000617_180 |
0.0726043187 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 7 |
Hb_002815_030 |
0.0727981796 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 8 |
Hb_002169_050 |
0.0735573688 |
- |
- |
PREDICTED: inositol 1,3,4-trisphosphate 5/6-kinase 4 [Jatropha curcas] |
| 9 |
Hb_012733_030 |
0.0739269759 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 10 |
Hb_000008_290 |
0.0739493831 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 11 |
Hb_005147_030 |
0.0762462579 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 12 |
Hb_003647_130 |
0.0771588578 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 13 |
Hb_002044_150 |
0.077946936 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 14 |
Hb_010174_040 |
0.0779505665 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 15 |
Hb_020378_030 |
0.0784397837 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 16 |
Hb_000614_240 |
0.0791401407 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 17 |
Hb_000053_040 |
0.0792774083 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 18 |
Hb_000130_160 |
0.07968361 |
- |
- |
PREDICTED: protein DJ-1 homolog B-like [Jatropha curcas] |
| 19 |
Hb_002471_290 |
0.080895311 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000207_180 |
0.0812386571 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |