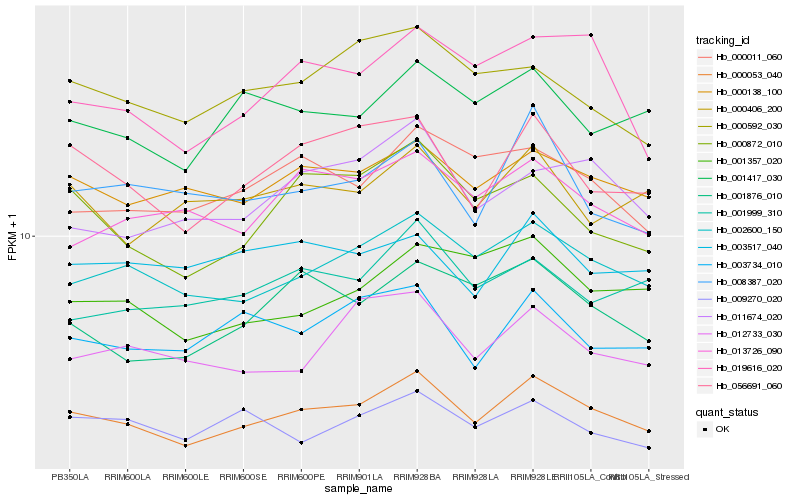
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000053_040 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 2 |
Hb_002600_150 |
0.0472786078 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000138_100 |
0.0546818604 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 4 |
Hb_056691_060 |
0.0592398992 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001417_030 |
0.0659431178 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 6 |
Hb_000011_060 |
0.0689483065 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003734_010 |
0.0697162079 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 8 |
Hb_000872_010 |
0.0711551345 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 9 |
Hb_000592_030 |
0.0716374468 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 10 |
Hb_001999_310 |
0.0718476382 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 11 |
Hb_009270_020 |
0.0718513421 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 12 |
Hb_013726_090 |
0.0720558955 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 13 |
Hb_012733_030 |
0.0721105389 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 14 |
Hb_000406_200 |
0.0722082019 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 15 |
Hb_001876_010 |
0.0723083621 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003517_040 |
0.0726783415 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001357_020 |
0.0727295011 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_019616_020 |
0.0728105069 |
- |
- |
PREDICTED: alanine--tRNA ligase [Jatropha curcas] |
| 19 |
Hb_011674_020 |
0.0739635388 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 20 |
Hb_008387_020 |
0.0740799607 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |