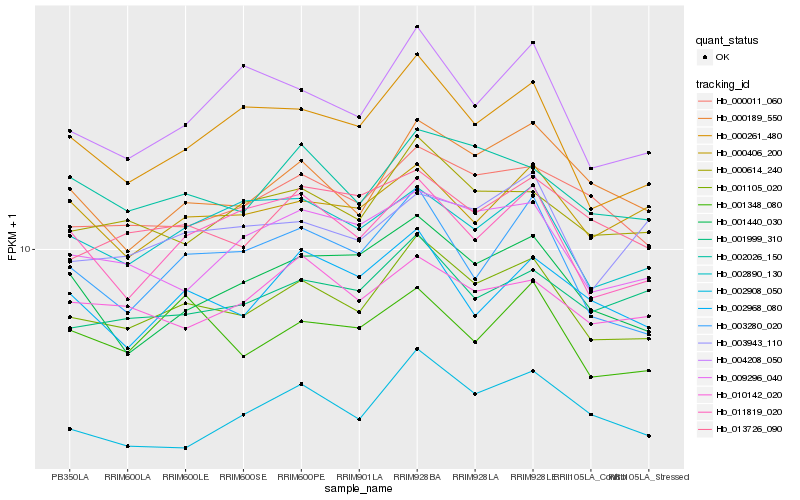
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001105_020 |
0.0 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 2 |
Hb_000614_240 |
0.0615176448 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 3 |
Hb_002026_150 |
0.0619318948 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair helicase XPB1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000261_480 |
0.0620287883 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 5 |
Hb_000011_060 |
0.0624185104 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000189_550 |
0.0659671768 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 7 |
Hb_009296_040 |
0.0697562836 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 8 |
Hb_002890_130 |
0.0698710571 |
- |
- |
tip120, putative [Ricinus communis] |
| 9 |
Hb_001999_310 |
0.0708452292 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 10 |
Hb_002908_050 |
0.0728264455 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 11 |
Hb_013726_090 |
0.0729082582 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 12 |
Hb_004208_050 |
0.0735969884 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 13 |
Hb_003280_020 |
0.0738413682 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011819_020 |
0.0744387106 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 15 |
Hb_010142_020 |
0.074687418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001348_080 |
0.0750351017 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 17 |
Hb_001440_030 |
0.0756179246 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 18 |
Hb_003943_110 |
0.0776440743 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 19 |
Hb_000406_200 |
0.0779236295 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 20 |
Hb_002968_080 |
0.0785766235 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |