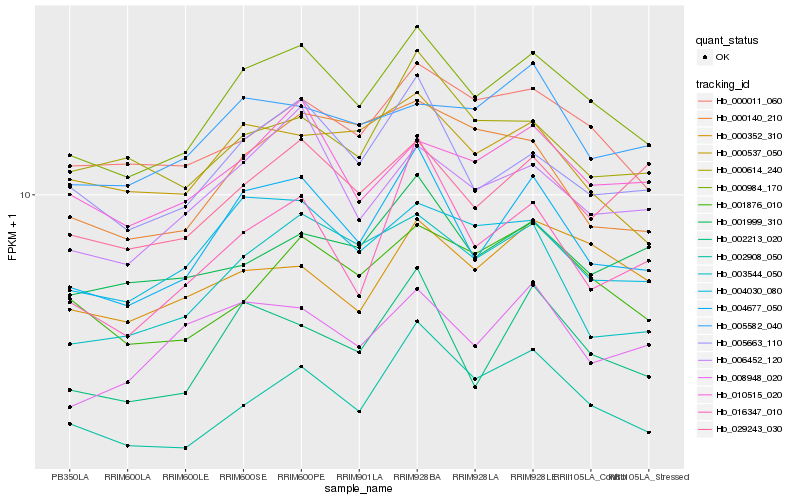
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000984_170 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 2 |
Hb_004677_050 |
0.0454573811 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004030_080 |
0.0558882919 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 4 |
Hb_005663_110 |
0.0593182627 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006452_120 |
0.0601907293 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 6 |
Hb_000011_060 |
0.0606813588 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002908_050 |
0.0608418564 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 8 |
Hb_001876_010 |
0.0633739265 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002213_020 |
0.0658411991 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010515_020 |
0.0659191001 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 11 |
Hb_003544_050 |
0.0707408535 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005582_040 |
0.0710561997 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 13 |
Hb_000537_050 |
0.0712379877 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 14 |
Hb_029243_030 |
0.0734195726 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000614_240 |
0.0746025989 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 16 |
Hb_000352_310 |
0.0757657626 |
- |
- |
hypothetical protein CISIN_1g0017891mg, partial [Citrus sinensis] |
| 17 |
Hb_000140_210 |
0.0770880086 |
- |
- |
sec10, putative [Ricinus communis] |
| 18 |
Hb_001999_310 |
0.0773160192 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 19 |
Hb_016347_010 |
0.0773935564 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 20 |
Hb_008948_020 |
0.0775224133 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |