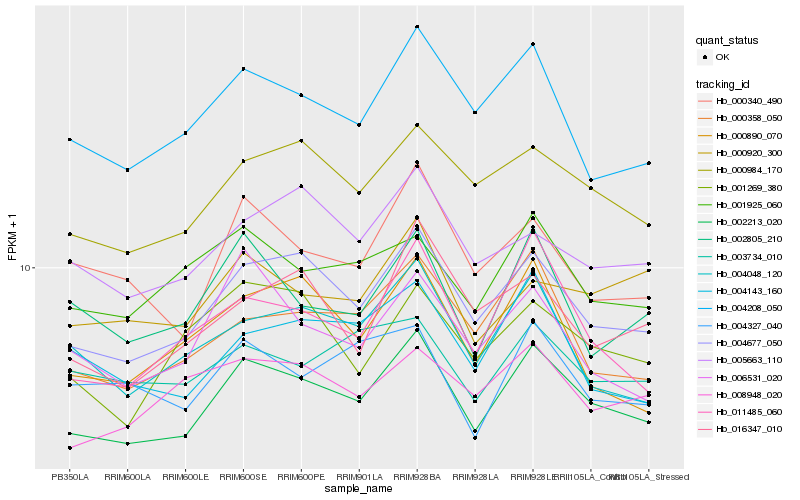
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002213_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004677_050 |
0.044575822 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000984_170 |
0.0658411991 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 4 |
Hb_004208_050 |
0.0731592279 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 5 |
Hb_011485_060 |
0.0751890141 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 6 |
Hb_000358_050 |
0.0781273094 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 7 |
Hb_001925_060 |
0.0802344924 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 8 |
Hb_000340_490 |
0.0810439532 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 9 |
Hb_004048_120 |
0.0814224941 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 10 |
Hb_002805_210 |
0.0831315192 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 11 |
Hb_005663_110 |
0.0850824459 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004143_160 |
0.0852162574 |
- |
- |
SAB, putative [Ricinus communis] |
| 13 |
Hb_006531_020 |
0.0867637532 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 14 |
Hb_008948_020 |
0.0870756654 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 15 |
Hb_001269_380 |
0.0879999256 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004327_040 |
0.0882135078 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 17 |
Hb_000920_300 |
0.0882718037 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 18 |
Hb_016347_010 |
0.0890546821 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 19 |
Hb_003734_010 |
0.0896278445 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 20 |
Hb_000890_070 |
0.089736377 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |