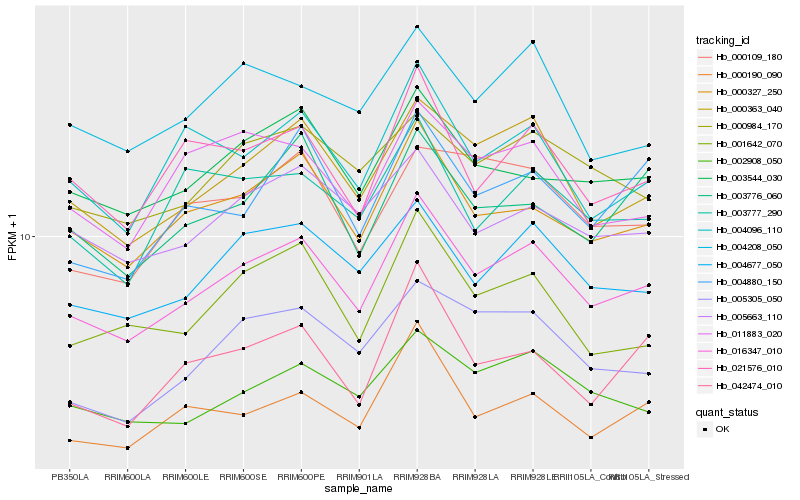
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016347_010 |
0.0 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 2 |
Hb_000363_040 |
0.0563177875 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 3 |
Hb_000327_250 |
0.0604737353 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 4 |
Hb_004677_050 |
0.0630267917 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002908_050 |
0.0719112718 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 6 |
Hb_003544_030 |
0.0731706792 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 7 |
Hb_021576_010 |
0.0738009337 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 8 |
Hb_005663_110 |
0.0769387383 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004096_110 |
0.0773798968 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 10 |
Hb_000984_170 |
0.0773935564 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_001642_070 |
0.0777473996 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 12 |
Hb_005305_050 |
0.0808377918 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 13 |
Hb_000190_090 |
0.0809815017 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 14 |
Hb_004208_050 |
0.0824173841 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 15 |
Hb_004880_150 |
0.082813377 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 16 |
Hb_011883_020 |
0.0832486475 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 17 |
Hb_003777_290 |
0.0843976861 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 18 |
Hb_003776_060 |
0.0868987843 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 19 |
Hb_000109_180 |
0.0871049896 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 20 |
Hb_042474_010 |
0.0876569428 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |