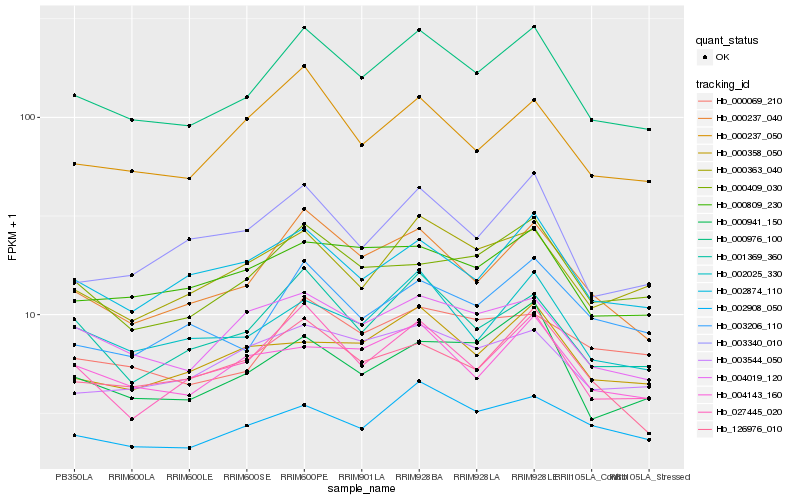
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_100 |
0.0 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 2 |
Hb_000237_040 |
0.0596401967 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 3 |
Hb_027445_020 |
0.0683360419 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 4 |
Hb_001369_360 |
0.0707058973 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 5 |
Hb_002025_330 |
0.0757407778 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 6 |
Hb_000363_040 |
0.0780869268 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 7 |
Hb_004019_120 |
0.0809086519 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 8 |
Hb_002874_110 |
0.0823880825 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 9 |
Hb_000237_050 |
0.0834074937 |
- |
- |
CP2 [Hevea brasiliensis] |
| 10 |
Hb_002908_050 |
0.0842199408 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 11 |
Hb_000941_150 |
0.0848881263 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000069_210 |
0.0849433142 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 13 |
Hb_000409_030 |
0.0854500118 |
- |
- |
PREDICTED: uncharacterized protein LOC105642693 [Jatropha curcas] |
| 14 |
Hb_004143_160 |
0.0858680347 |
- |
- |
SAB, putative [Ricinus communis] |
| 15 |
Hb_003340_010 |
0.0859324869 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 16 |
Hb_003206_110 |
0.0862919044 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 17 |
Hb_003544_050 |
0.0863829837 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000809_230 |
0.0868045428 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 19 |
Hb_126976_010 |
0.0890757887 |
- |
- |
- |
| 20 |
Hb_000358_050 |
0.0901143315 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |