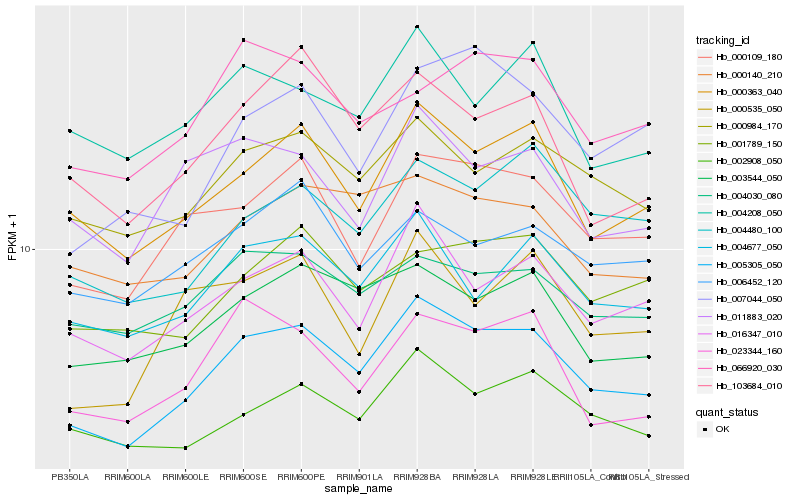
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005305_050 |
0.0 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 2 |
Hb_000363_040 |
0.0730396983 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 3 |
Hb_000109_180 |
0.078921165 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 4 |
Hb_016347_010 |
0.0808377918 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 5 |
Hb_000984_170 |
0.0841075057 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 6 |
Hb_003544_050 |
0.0862132027 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002908_050 |
0.0879217714 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 8 |
Hb_004677_050 |
0.088020186 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004480_100 |
0.0926608211 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 10 |
Hb_000140_210 |
0.0928640391 |
- |
- |
sec10, putative [Ricinus communis] |
| 11 |
Hb_103684_010 |
0.0940400384 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 12 |
Hb_023344_160 |
0.0948949774 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 13 |
Hb_000535_050 |
0.0983071864 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 14 |
Hb_004030_080 |
0.0984387288 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 15 |
Hb_007044_050 |
0.0994510616 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 16 |
Hb_001789_150 |
0.0997565395 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011883_020 |
0.100339404 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 18 |
Hb_004208_050 |
0.102699783 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 19 |
Hb_066920_030 |
0.1037258549 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 20 |
Hb_006452_120 |
0.1040171262 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |