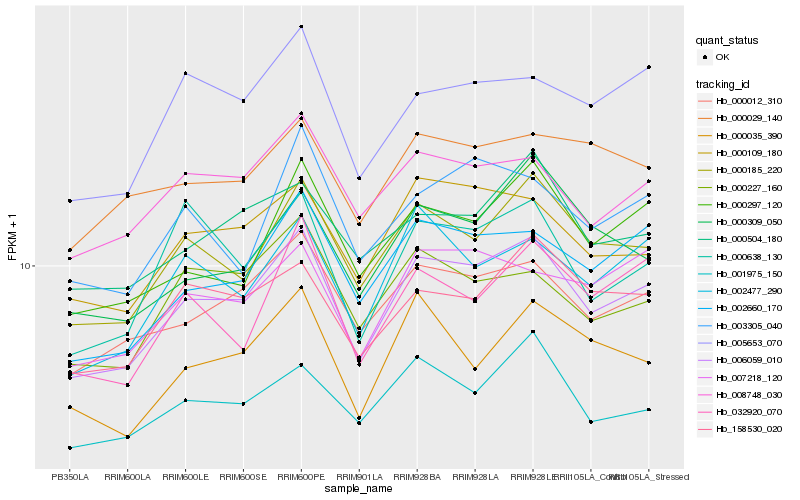
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006059_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 2 |
Hb_000185_220 |
0.0630738418 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 3 |
Hb_000012_310 |
0.063077052 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_002660_170 |
0.069455339 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_000109_180 |
0.0737024208 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 6 |
Hb_008748_030 |
0.0776495365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000297_120 |
0.0779817902 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 8 |
Hb_158530_020 |
0.0808057019 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_002477_290 |
0.0818158391 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_001975_150 |
0.0821932485 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 11 |
Hb_007218_120 |
0.0828840283 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 12 |
Hb_005653_070 |
0.0841126834 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 13 |
Hb_000504_180 |
0.0853245695 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 14 |
Hb_000227_160 |
0.0864399034 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 15 |
Hb_000638_130 |
0.0869978711 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 16 |
Hb_032920_070 |
0.0870592706 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 17 |
Hb_000309_050 |
0.0878934121 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003305_040 |
0.0895134202 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 19 |
Hb_000029_140 |
0.0910924466 |
- |
- |
PREDICTED: putative GDP-L-fucose synthase 2 [Populus euphratica] |
| 20 |
Hb_000035_390 |
0.0916200086 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |