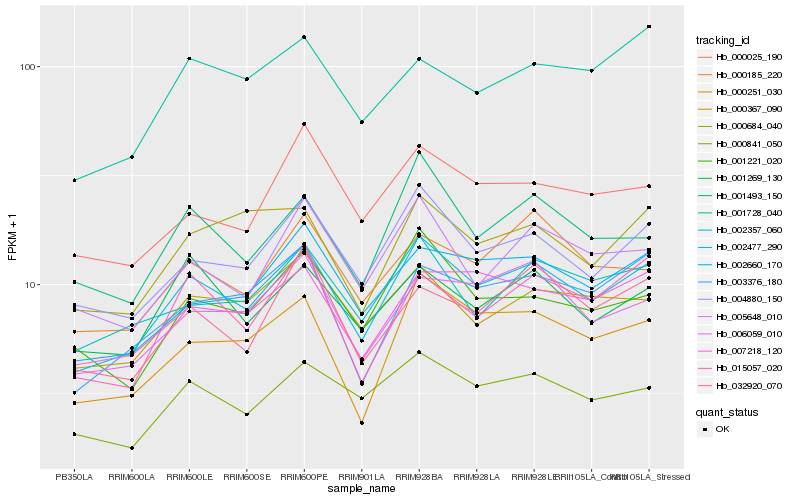
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_290 |
0.0 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_004880_150 |
0.0650918639 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 3 |
Hb_000841_050 |
0.0762846633 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 4 |
Hb_002660_170 |
0.078330372 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_007218_120 |
0.0787358318 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000251_030 |
0.0787449832 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_006059_010 |
0.0818158391 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 8 |
Hb_002357_060 |
0.0837306956 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 9 |
Hb_000367_090 |
0.0843081703 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 10 |
Hb_003376_180 |
0.0851889341 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 11 |
Hb_015057_020 |
0.0856905459 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 12 |
Hb_001221_020 |
0.0868247701 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 13 |
Hb_001493_150 |
0.0868637635 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 14 |
Hb_005648_010 |
0.0869491395 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 15 |
Hb_001269_130 |
0.0870687621 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 16 |
Hb_000185_220 |
0.0892255813 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 17 |
Hb_032920_070 |
0.0892271115 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 18 |
Hb_000684_040 |
0.0899855733 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 19 |
Hb_001728_040 |
0.0916176408 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 20 |
Hb_000025_190 |
0.0918186206 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |