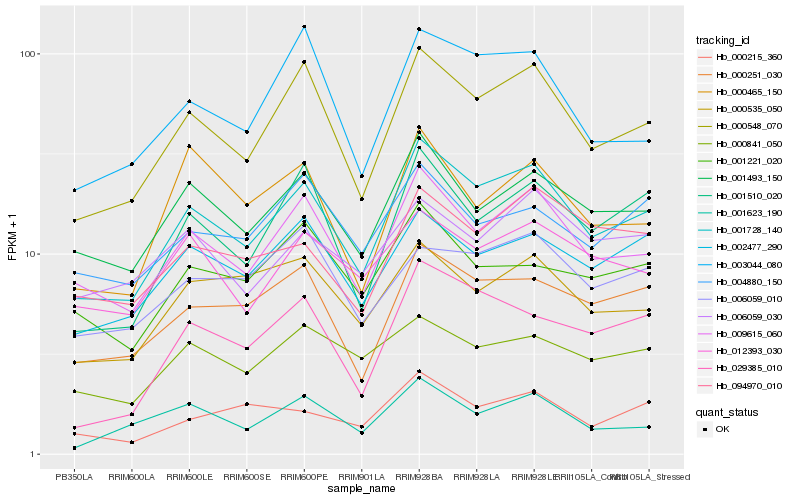
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001728_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_000548_070 |
0.0557931299 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_009615_060 |
0.0675193433 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 4 |
Hb_001493_150 |
0.0875745833 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 5 |
Hb_000251_030 |
0.091375811 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001510_020 |
0.0954619936 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 7 |
Hb_002477_290 |
0.098524664 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_094970_010 |
0.1085128764 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003044_080 |
0.1090669675 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_000535_050 |
0.1097863211 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 11 |
Hb_029385_010 |
0.1115671626 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 12 |
Hb_012393_030 |
0.1123898669 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 13 |
Hb_000841_050 |
0.1153678256 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 14 |
Hb_000215_360 |
0.1157281835 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_006059_030 |
0.117254479 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_006059_010 |
0.1174862233 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 17 |
Hb_004880_150 |
0.1178038393 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 18 |
Hb_001221_020 |
0.1179408557 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 19 |
Hb_001623_190 |
0.1196374241 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 20 |
Hb_000465_150 |
0.120091898 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |