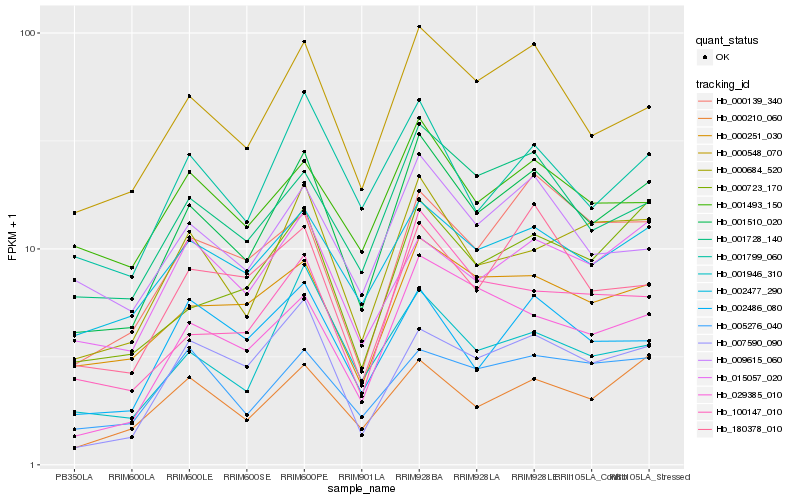
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001510_020 |
0.0 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 2 |
Hb_000548_070 |
0.0812683871 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_001728_140 |
0.0954619936 |
- |
- |
- |
| 4 |
Hb_002477_290 |
0.0985150617 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000684_520 |
0.0999520471 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 6 |
Hb_000210_060 |
0.104013331 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 7 |
Hb_000251_030 |
0.1046257984 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_001799_060 |
0.1081596065 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 9 |
Hb_000723_170 |
0.1087575857 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 10 |
Hb_029385_010 |
0.1120018679 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 11 |
Hb_009615_060 |
0.1145996434 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 12 |
Hb_007590_090 |
0.1146336015 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 13 |
Hb_001946_310 |
0.1156339276 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 14 |
Hb_100147_010 |
0.116072372 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 15 |
Hb_015057_020 |
0.1166092146 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 16 |
Hb_000139_340 |
0.1170962342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_180378_010 |
0.1205410534 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 18 |
Hb_001493_150 |
0.1214280757 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 19 |
Hb_002486_080 |
0.1227081582 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 20 |
Hb_005276_040 |
0.1230431607 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |