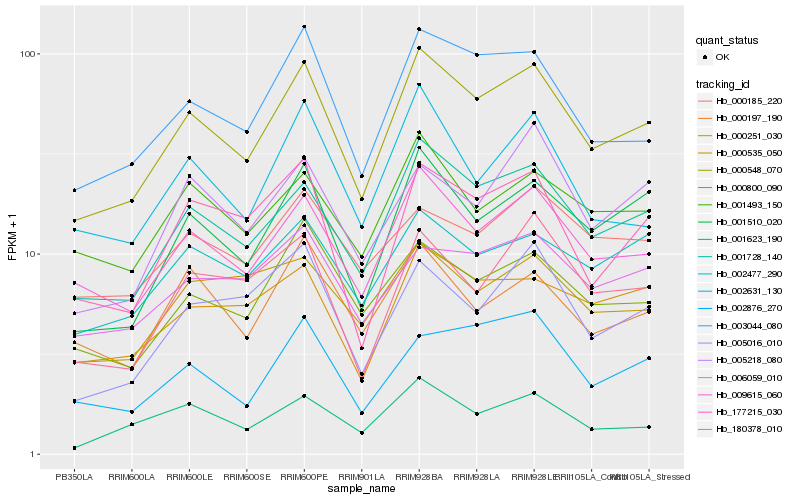
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000548_070 |
0.0 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 2 |
Hb_001728_140 |
0.0557931299 |
- |
- |
- |
| 3 |
Hb_009615_060 |
0.0654817236 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 4 |
Hb_003044_080 |
0.0779328123 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_001510_020 |
0.0812683871 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 6 |
Hb_000800_090 |
0.0933784532 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000251_030 |
0.0987335092 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_177215_030 |
0.0987356441 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_002477_290 |
0.0997418568 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_001493_150 |
0.1011032329 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 11 |
Hb_001623_190 |
0.103466577 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 12 |
Hb_180378_010 |
0.1041601092 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 13 |
Hb_006059_010 |
0.1060175179 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 14 |
Hb_002631_130 |
0.1070528087 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 15 |
Hb_002876_270 |
0.1076104878 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000197_190 |
0.1093551731 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_005016_010 |
0.1095885434 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 18 |
Hb_000185_220 |
0.1107922482 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 19 |
Hb_005218_080 |
0.1120460876 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000535_050 |
0.1138883223 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |