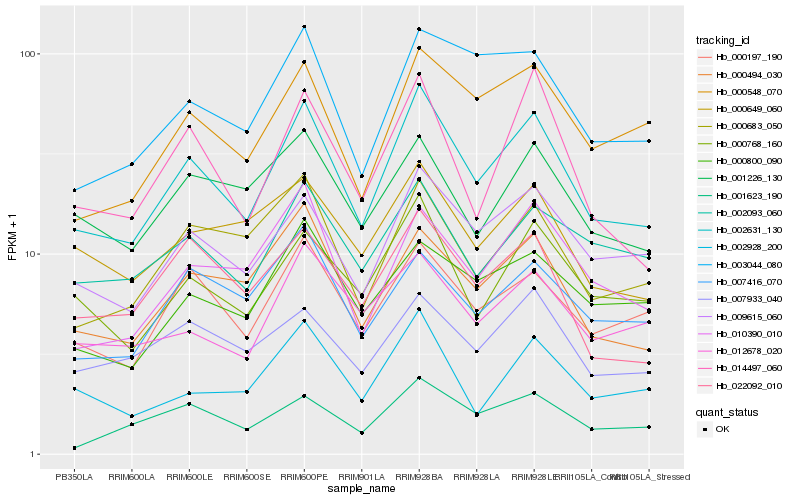
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_130 |
0.0 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 2 |
Hb_009615_060 |
0.0882648465 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 3 |
Hb_000197_190 |
0.0984178309 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000494_030 |
0.0994030077 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 5 |
Hb_000768_160 |
0.1009431456 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 6 |
Hb_000683_050 |
0.1051984015 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 7 |
Hb_000548_070 |
0.1070528087 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_010390_010 |
0.1085346565 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 9 |
Hb_014497_060 |
0.1108039085 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 10 |
Hb_003044_080 |
0.1120141764 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 11 |
Hb_022092_010 |
0.1120149651 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 12 |
Hb_007933_040 |
0.1156198494 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002093_060 |
0.1157779419 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 14 |
Hb_001226_130 |
0.1175795585 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 15 |
Hb_002928_200 |
0.1179652816 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 16 |
Hb_000649_060 |
0.1180014333 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 17 |
Hb_007416_070 |
0.1183978333 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 18 |
Hb_000800_090 |
0.1186703872 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001623_190 |
0.1186876405 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 20 |
Hb_012678_020 |
0.1192707104 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |