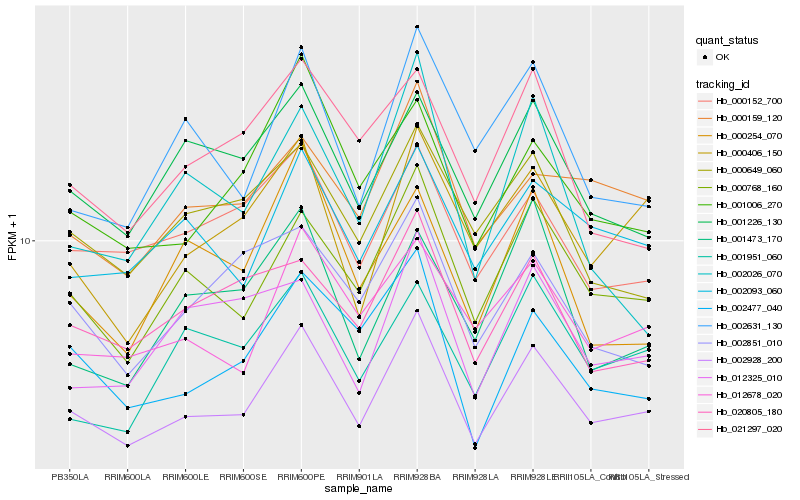
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002928_200 |
0.0 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 2 |
Hb_000768_160 |
0.0877371338 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 3 |
Hb_000406_150 |
0.1145896821 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 4 |
Hb_002631_130 |
0.1179652816 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 5 |
Hb_002477_040 |
0.1194632929 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_012678_020 |
0.1209792762 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001006_270 |
0.1248565836 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 8 |
Hb_002093_060 |
0.1264954782 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_001226_130 |
0.1294308893 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 10 |
Hb_000152_700 |
0.1313895644 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 11 |
Hb_000649_060 |
0.1337354235 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 12 |
Hb_001951_060 |
0.1338945771 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 13 |
Hb_020805_180 |
0.1341558597 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 14 |
Hb_000159_120 |
0.1345947467 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 15 |
Hb_000254_070 |
0.1346368724 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 16 |
Hb_002851_010 |
0.1366160287 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_021297_020 |
0.1368268382 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 18 |
Hb_002026_070 |
0.1368591578 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 19 |
Hb_012325_010 |
0.1380818579 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 20 |
Hb_001473_170 |
0.1393720384 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |