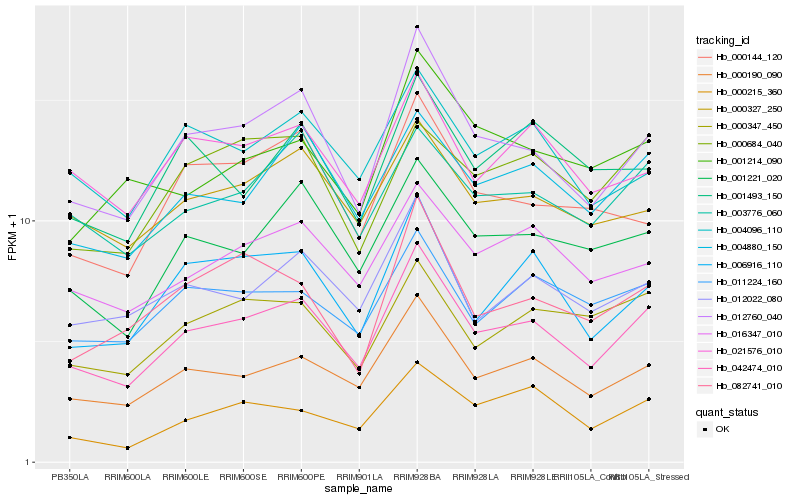
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042474_010 |
0.0 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 2 |
Hb_012760_040 |
0.0553162543 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 3 |
Hb_000190_090 |
0.0593293942 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 4 |
Hb_004880_150 |
0.0831820835 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 5 |
Hb_001221_020 |
0.0867935766 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 6 |
Hb_006916_110 |
0.0874954922 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_016347_010 |
0.0876569428 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 8 |
Hb_012022_080 |
0.0876649993 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_003776_060 |
0.092383858 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 10 |
Hb_000327_250 |
0.0935976444 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 11 |
Hb_011224_160 |
0.0969612433 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 12 |
Hb_021576_010 |
0.0978480477 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 13 |
Hb_004096_110 |
0.0988336303 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 14 |
Hb_000347_450 |
0.1001198133 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000215_360 |
0.1013757429 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 16 |
Hb_082741_010 |
0.1030903326 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 17 |
Hb_000684_040 |
0.1068546041 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 18 |
Hb_000144_120 |
0.1072219653 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_001493_150 |
0.1104346649 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 20 |
Hb_001214_090 |
0.1113330017 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |