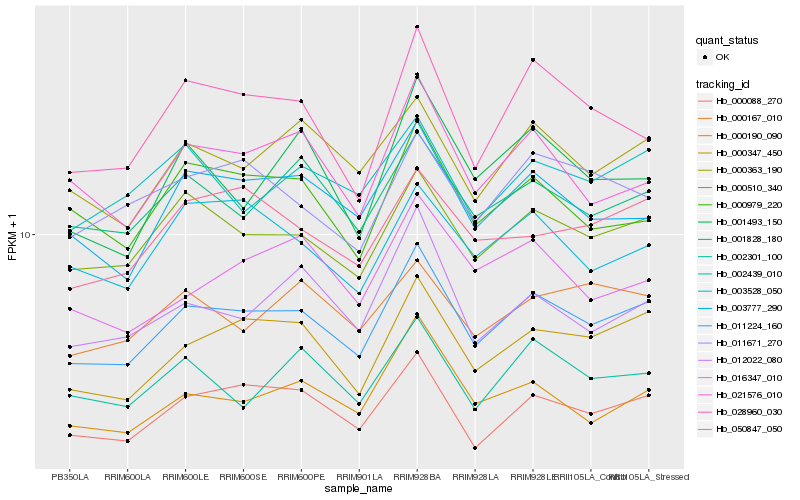
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011224_160 |
0.0 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 2 |
Hb_000510_340 |
0.0596249359 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 3 |
Hb_000347_450 |
0.0621194274 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000088_270 |
0.0674463203 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 5 |
Hb_003777_290 |
0.0710245692 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 6 |
Hb_000363_190 |
0.074306868 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_021576_010 |
0.077383593 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 8 |
Hb_001828_180 |
0.0784140351 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 9 |
Hb_028960_030 |
0.0790378723 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 10 |
Hb_000190_090 |
0.0796213259 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 11 |
Hb_000979_220 |
0.0844740697 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 12 |
Hb_050847_050 |
0.0846309987 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012022_080 |
0.0848304566 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_002439_010 |
0.086883714 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 15 |
Hb_003528_050 |
0.0879430837 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 16 |
Hb_011671_270 |
0.0883117142 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 17 |
Hb_016347_010 |
0.0884435567 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 18 |
Hb_002301_100 |
0.0901629879 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 19 |
Hb_001493_150 |
0.090326995 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 20 |
Hb_000167_010 |
0.0914292537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |