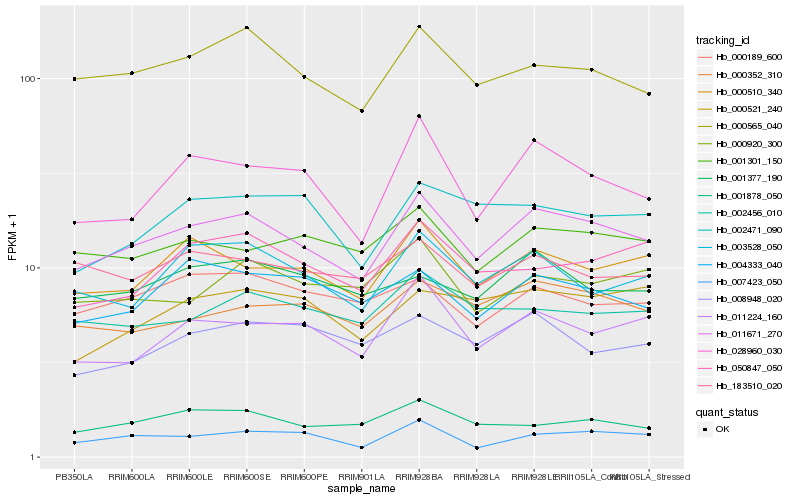
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_270 |
0.0 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 2 |
Hb_000565_040 |
0.0772065898 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_000510_340 |
0.0790714457 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 4 |
Hb_028960_030 |
0.0847080008 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 5 |
Hb_003528_050 |
0.086021315 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 6 |
Hb_000352_310 |
0.0881031881 |
- |
- |
hypothetical protein CISIN_1g0017891mg, partial [Citrus sinensis] |
| 7 |
Hb_011224_160 |
0.0883117142 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 8 |
Hb_007423_050 |
0.0911008025 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 3 [Jatropha curcas] |
| 9 |
Hb_001301_150 |
0.0925810781 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 10 |
Hb_004333_040 |
0.0954962834 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 11 |
Hb_050847_050 |
0.0957636304 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002456_010 |
0.0961178392 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_000920_300 |
0.0980894641 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 14 |
Hb_001878_050 |
0.0982300026 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 15 |
Hb_002471_090 |
0.0985784791 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 16 |
Hb_000189_600 |
0.0987274943 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_183510_020 |
0.0996424411 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 18 |
Hb_001377_190 |
0.1002315497 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_008948_020 |
0.102295163 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 20 |
Hb_000521_240 |
0.1024606498 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |