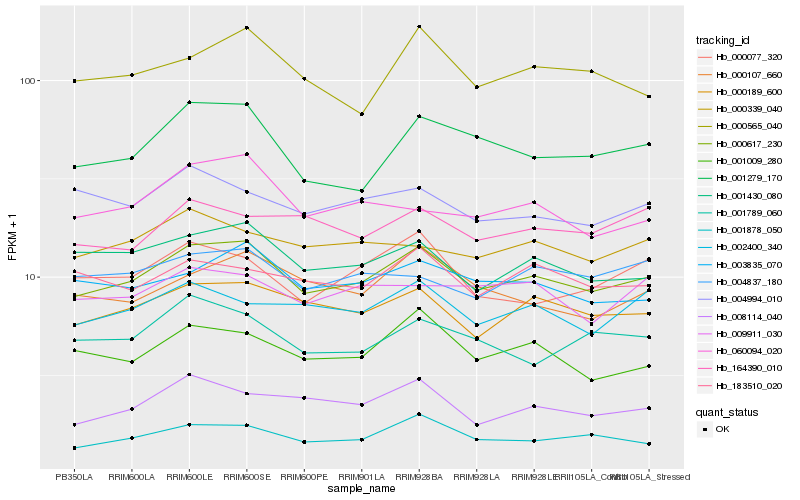
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_230 |
0.0 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 2 |
Hb_000189_600 |
0.0589167754 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_001430_080 |
0.0663711104 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 4 |
Hb_001878_050 |
0.066550543 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 5 |
Hb_003835_070 |
0.0689749049 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001279_170 |
0.0712658511 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_001009_280 |
0.0729031812 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 8 |
Hb_008114_040 |
0.0758358378 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 9 |
Hb_000107_660 |
0.0759373093 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 10 |
Hb_002400_340 |
0.0762722295 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 11 |
Hb_060094_020 |
0.077501917 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000339_040 |
0.0782063263 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_183510_020 |
0.0786364264 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 14 |
Hb_000565_040 |
0.0790247785 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 15 |
Hb_004837_180 |
0.0795220938 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000077_320 |
0.0795668086 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 17 |
Hb_001789_060 |
0.0798371982 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 18 |
Hb_009911_030 |
0.0812545655 |
transcription factor |
TF Family: FAR1 |
far-red impaired response 1 -like protein [Gossypium arboreum] |
| 19 |
Hb_004994_010 |
0.0813994851 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 20 |
Hb_164390_010 |
0.081720119 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |