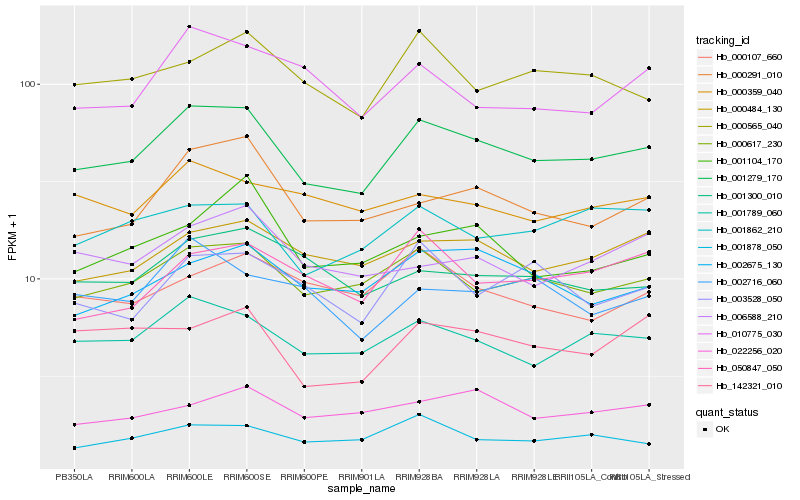
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_170 |
0.0 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000617_230 |
0.0712658511 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 3 |
Hb_001789_060 |
0.0713369682 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 4 |
Hb_000484_130 |
0.0875369471 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 5 |
Hb_000291_010 |
0.0880412099 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 6 |
Hb_001862_210 |
0.0912002501 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: salutaridine reductase-like [Citrus sinensis] |
| 7 |
Hb_002675_130 |
0.0923419438 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 16, NatA auxiliary subunit isoform X1 [Jatropha curcas] |
| 8 |
Hb_001878_050 |
0.092978531 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000565_040 |
0.0941710955 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_003528_050 |
0.0992391289 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 11 |
Hb_001300_010 |
0.099717851 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_022256_020 |
0.099814706 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35130, chloroplastic [Jatropha curcas] |
| 13 |
Hb_050847_050 |
0.10105077 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 14 |
Hb_142321_010 |
0.1010700201 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 15 |
Hb_010775_030 |
0.1017805346 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 16 |
Hb_001104_170 |
0.1023677444 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 17 |
Hb_000359_040 |
0.1024131444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006588_210 |
0.1029651689 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 19 |
Hb_002716_060 |
0.1042088341 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 20 |
Hb_000107_660 |
0.1051745808 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |