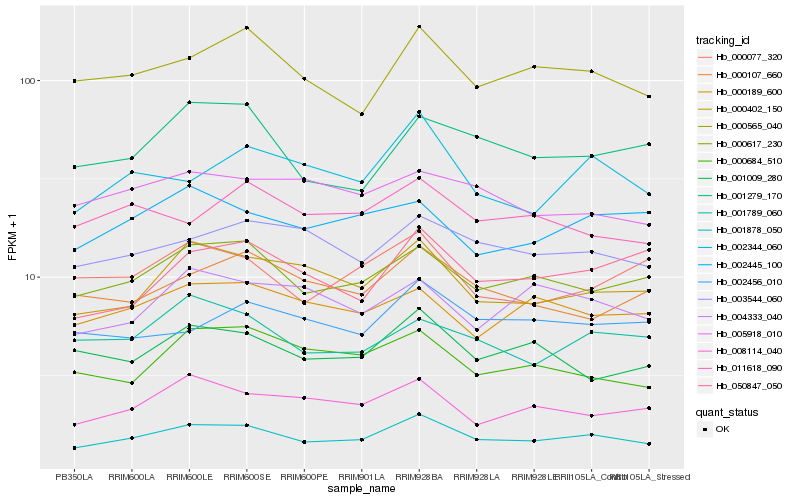
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001878_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000617_230 |
0.066550543 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 3 |
Hb_000402_150 |
0.0760283075 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000565_040 |
0.0762204489 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_003544_060 |
0.0813846418 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 6 |
Hb_008114_040 |
0.0857636986 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 7 |
Hb_002344_060 |
0.086836177 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 8 |
Hb_001009_280 |
0.0887081156 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 9 |
Hb_000189_600 |
0.0905345805 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_000107_660 |
0.0910043642 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 11 |
Hb_001789_060 |
0.0915879225 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 12 |
Hb_002456_010 |
0.0922106773 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_002445_100 |
0.0926974643 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001279_170 |
0.092978531 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000684_510 |
0.0938989875 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 16 |
Hb_000077_320 |
0.093938036 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 17 |
Hb_050847_050 |
0.0939540997 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011618_090 |
0.0940390239 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_005918_010 |
0.0952281965 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 20 |
Hb_004333_040 |
0.0967642136 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |