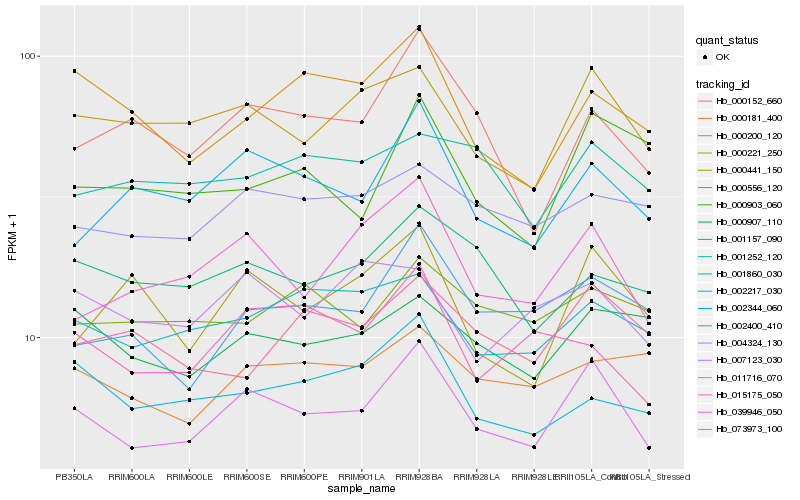
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011716_070 |
0.0 |
transcription factor |
TF Family: HRT |
PREDICTED: uncharacterized protein LOC105637858 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000221_250 |
0.0817438011 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 3 |
Hb_015175_050 |
0.0910852307 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 4 |
Hb_002344_060 |
0.0949699453 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 5 |
Hb_001860_030 |
0.0980742967 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 6 |
Hb_002400_410 |
0.0983877557 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 7 |
Hb_004324_130 |
0.099847391 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 8 |
Hb_000907_110 |
0.1021530442 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 9 |
Hb_000152_660 |
0.1026490814 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 10 |
Hb_001157_090 |
0.1052504835 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 11 |
Hb_002217_030 |
0.1065534183 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000181_400 |
0.1076609967 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000200_120 |
0.1085352449 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000441_150 |
0.1090869975 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000556_120 |
0.1093062792 |
- |
- |
Spastin, putative [Ricinus communis] |
| 16 |
Hb_001252_120 |
0.1093459401 |
- |
- |
PREDICTED: delta-1-pyrroline-5-carboxylate synthase-like [Gossypium raimondii] |
| 17 |
Hb_039946_050 |
0.1099185479 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_000903_060 |
0.1100256138 |
- |
- |
PREDICTED: ras-related protein RABA5e [Jatropha curcas] |
| 19 |
Hb_007123_030 |
0.1100721563 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 20 |
Hb_073973_100 |
0.1104677649 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |